Abstract
Neglected tropical disease drug discovery requires application of pragmatic and efficient methods for development of new therapeutic agents. In this report we describe our target repurposing efforts for the essential phosphodiesterase (PDE) enzymes TbrPDEB1 and TbrPDEB2 of Trypanosoma brucei, the causative agent for human African trypanosomiasis (HAT). We describe protein expression and purification, assay development, and benchmark screening of a collection of 20 established human PDE inhibitors. We disclose that the human PDE4 inhibitor piclamilast, and some of its analogs, show modest inhibition of TbrPDEB1 and B2, and quickly kill the bloodstream form of the subspecies T. brucei brucei. We also report the development of a homology model of TbrPDEB1 that is useful for understanding the compound-enzyme interactions and for comparing the parasitic and human enzymes. Our profiling and early medicinal chemistry results strongly suggest that human PDE4 chemotypes represent a better starting point for optimization of TbrPDEB inhibitors than those that target any other human PDEs.
Introduction
Human African trypanosomiasis (HAT), or African sleeping sickness, is a neglected tropical disease that affects tens of thousands of patients annually.1 The disease is fatal unless treated and current treatments are limited in both efficacy and safety.2 Financial incentives for drug discovery against HAT are quite limited due to the economically disadvantaged regions where this disease is endemic. As a strategy to overcome this disincentive for drug discovery, we hypothesized that medicinal chemistry knowledge against human drug targets could be repurposed to facilitate rapid and cost-effective drug discovery against parasite drug targets.3 In this approach, existing drugs and drug-like compounds are used to pharmacologically validate parasite targets, and to serve as early hits or leads from which to optimize parasite-specific therapeutics.
Nucleotide phosphodiesterases (PDEs) are vital regulators of signalling pathways, hydrolyzing the secondary messengers cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP). Humans have 11 families of PDEs, most of which have numerous isoforms. PDEs are involved in a variety of biological processes and have been implicated in a wide range of therapeutic indications.4-7 Despite the size of the PDE family and active site sequence identity between isoforms ranging from 25-52%, highly selective inhibitors have been developed, leading to clinical candidates and drugs for chronic obstructive pulmonary disorder (PDE4), erectile dysfunction (PDE5) and schizophrenia (PDE10).
By contrast, Trypanosoma brucei has only five PDEs, all of which are specific for cAMP.8 The Trypanosoma brucei PDEB (TbrPDEB) family is encoded by two tandemly arranged genes and its members have distinct sub-cellular localizations.9 TbrPDEB1 and TbrPDEB2 are not individually essential for parasite survival but when the enzymes are knocked down by RNAi simultaneously, the parasites are incapable of proper cell division and ultimately die.9,10
In this report we describe the selection of TbrPDEB1 and TbrPDEB2 from a list of prioritized drug targets from the T. brucei genome and our initial inhibitor chemotype explorations. In confirmation of the previous RNAi experiments described above, these targets are validated pharmacologically in both in vitro biochemical assays and in parasite cultures. An initial survey of chemical space shows tolerance of structural modifications of the lead chemotype in regions that can potentially be exploited to optimize selectivity and potency.
Results
The Trypanosoma brucei brucei genome was examined for the presence of homologs of known, chemically validated drug targets that have compounds that have successfully passed Phase II clinical trials. Since the pathogens infiltrate the central nervous system (CNS), we further restricted our focus to parasite homologs of known targets with precedents for clinical compounds capable of crossing the blood brain barrier, from which we selected the T. brucei brucei cyclic AMP phosphodiesterases TbrPDEB1 (Tb09.160.3590) and TbrPDEB2 (Tb09.160.3630). The catalytic domains of the trypanosomal PDEB enzymes have 30-35% identity to the successful PDE targets from humans, and others have demonstrated that certain established human PDE inhibitors are active against partially purified preparations of TbrPDEB1and B2.11,12 We therefore hypothesized that a broader exploration of human PDE inhibitor chemotypes could identify good starting points for chemical optimization. A similar approach was recently reported, describing the profiling of human PDE5 inhibitors as a starting point for antimalarial agents.13
The His-tagged catalytic domains of TbrPDEB1 (Ser580-Arg930) and TbrPDEB2 (Ser580-Ser925) were expressed using the Sf21/baculovirus system and purified by nickel-sepharose chromatography, yielding milligram quantities of active protein. The kinetic parameters of the recombinant catalytic domains were determined, and the KM of cAMP with the TbrPDEB1 was measured to be 40 μM (±3.4), and with the TbrPDEB2 catalytic domain the KM was 11.5 μM (±4.4). Specific activities of 0.35 nmol/min/μg (±0.01) and 0.12 nmol/min/μg (±0.01) were observed for TbrPDEB1 and TbrPDEB2 respectively. Thus, while both proteins behaved similarly biochemically, TbrPDEB2 had a slightly lower KM and specific activity compared with TbrPDEB1. The KM of the catalytic domain of TbrPDEB2 is lower than that reported previously (KM ≥ 44 IM) and was more similar to the full length protein with the cAMP binding GAF domains intact (4.5 IM).14 The inverse is seen with TbrPDEB1 where the previously described KM of 8 IM is significantly lower than that observed with our TbrPDEB1.12 However, in previous reports, TbrPDEB1 and TbrPDEB2 were expressed as full-length enzymes, in different expression systems (yeast and human cells respectively), and neither were purified; these differences may explain the disparity in observed KM values, and we therefore judged these differences acceptable and our proteins suitable for supporting inhibitor development.
Extensive sets of human PDE inhibitors exist, and these compounds represent potential starting points for new inhibitor discovery against these trypanosomal PDEs. Our profiling set (annotated in the Supporting Information) includes a representative sampling of PDE inhibitor chemotypes that have been disclosed to date. Consistent with our preference to build on scaffolds with evidence for safety and tolerability, wherever possible we included compounds that have been reported in advanced stages of preclinical development or which have been approved for use. While the T. brucei PDEBs are known to be cAMP-selective, both approved drugs and development candidates against cGMP-specific PDEs (such as PDE5) were included, as were development candidates against those that hydrolyze both cAMP and cGMP (such as PDE10). Compounds were selected and synthesized using published methods, donated, or purchased, and were tested at a single concentration.
We sought to identify compounds capable of inhibiting both TbrPDEB1 and TbrPDEB2 since RNAi only kills trypanosomes when both enzymes are disrupted. The feasibility of achieving dual inhibition is supported by analysis of the protein sequences. Overall protein sequence identity between TbrPDEB1 and TbrPDEB2 is 75% using a ClustalW2 alignment,15 and the sequence identity within the catalytic domains of these two proteins is 88%. The 3D structure alignment of TbrPDEB1, TbrPDEB2, human PDE4 and L. major PDEB1 is shown in the Supporting Information in tabular form; residues involved in binding are highlighted. The high similarity between the active sites of TbrPDEB1 and TbrPDEB2 suggests that identification of a compound that inhibits both TbrPDEB1 and B2 should be an achievable goal. We elected to perform primary screening against TbrPDEB1, followed by secondary screening against TbrPDEB2 with inhibitors that show better than 20 μM inhibitory potency (IC50) against TbrPDEB1.
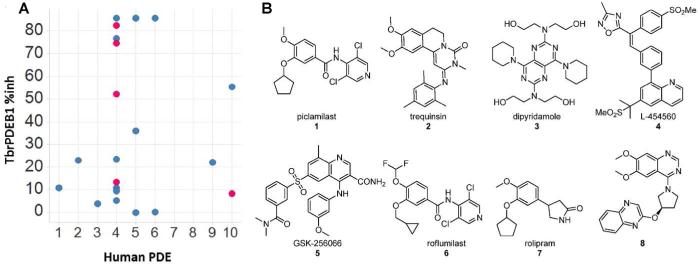
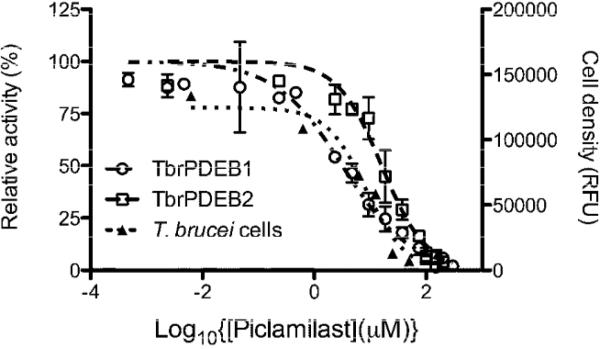
The scatter plot in Figure 1A shows the percent inhibition data as a function of human PDE inhibitor class, and Table 1 contains dose-response assessments of those inhibitors showing greater than 65% inhibition at the single dose concentration of benchmarked inhibitors. Initial tests of the PDE4 inhibitors piclamilast (1) and trequinsin (2), and the PDE4/6 inhibitor dipyridamole (3) (the latter two tested at 100 μM) showed over 50% inhibition. Subsequent tests of additional PDE4 inhibitors (L-454560 (4) and GSK-256066, (5) also inhibited TbrPDEB1 by at least 50% at 10 μM. Assessment of 1 in dose-response studies revealed IC50 values of 4.7 μM against TbrPDEB1, and 11.4 μM versus TbrPDEB2 (Figure 2). While 1 demonstrated the greatest potency against TbrPDEB1 among the compounds tested, the closely-related PDE4 inhibitor roflumilast (6) was essentially inactive, as was rolipram (7), a compound sharing a substantial substructure with piclamilast. Compound 8, a recently-disclosed human PDE10 inhibitor,16 displayed 55% inhibition at 100 μM. Recognizing the extensive precedence in human PDE4 inhibitors,17-19 including a number of compounds that have entered clinical trials, we made this family of inhibitors our primary focus.
Figure 1.
(A) Benchmark screening data of human PDE inhibitors against TbrPDEB1 at 100 μM (red) and 10 μM (blue). Tabulation of the experimental data is contained in the Supporting Information; (B) Representative PDE inhibitors tested in Figure 1A.
Table 1.
Summary of dose-response of compounds showing >65% inhibition against TbrPDEB1.
| Compound | hPDE | TbrPDEB1 IC50 (μM)a | TbrPDEB2 IC50 (μM)a | Cell culture (EC50, μM) |
|---|---|---|---|---|
| Dipyridamole (3) | ~72 ± 10b | nt | nt | |
| L-454560 (4) | 4 | 8.8 ± 1.9 | 3.9 ±0.8 | 10.8 ± 8.9 |
| Piclamilast (1) | 4 | 4.7 ± 1.0 | 11.4± 1.1 | 9.57 ± 0.9 |
| Roflumilast (6) | 4 | >> 100 | nt | nt |
| Rolipram (7) | 4 | >>100 | nt | nt |
| Trequinsin (2) | 4,5,6 | 12.0 ± 1.4 | 25.4 ±2.6 | 5.8 ± 0.4 |
| BAY 19-800431 | 4 | >>10 | >>10 | nt |
| RO 20-172432 | 4 | >>100 | nt | nt |
IC50 values reported to be “>> x μM” if <25% inh at x μM; “> x μM” if 25-50% inh at x μM.
Measurement of a precise IC50 value was precluded by low compound solubility.
Figure 2.
Dose-response curves for piclamilast against TbrPDEB1 (circles), TbrPDEB2 (squares) and bloodstream form T. brucei brucei (triangles).
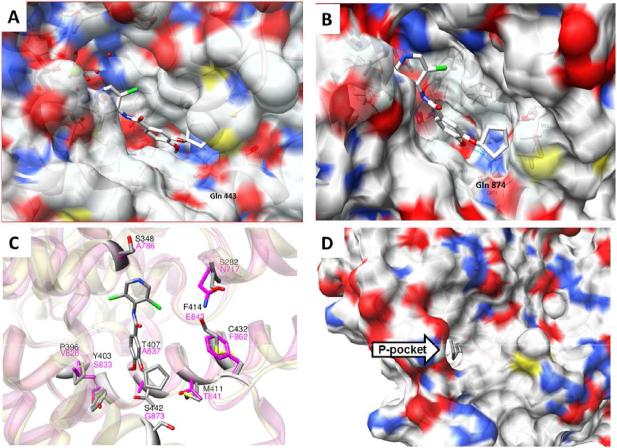
To help elucidate binding site features that could drive potency differences between the parasite and human enzymes, and to facilitate compound binding hypotheses to drive medicinal chemistry optimization, homology models of TbrPDEB1 and B2 were constructed. Multiple template structures were tried, but the Leishmania major PDEB1 (LmjPDEB1) crystal structure20 yielded the best-scoring model structures and is most closely related to our target (Supplementary Material). Figure 3A shows the X-ray crystal structure previously reported for piclamilast bound to hPDE4 (PDB ID 1XM4),21 and the predicted pose for 1 in the model structure of TbrPDEB1 (Figure 3B). A superposition of the TbrPDEB1 model structure (pink) with the human PDE4 crystal structure (gray) is shown in Figure 3C, highlighting the modest amino acid differences in the proximity of the drug binding site that are likely implicated in the differential binding potency between the human and parasite enzymes.
Figure 3.
(A) The crystal structure of human PDE4B complexes with piclamilast (PDB ID: 1XM4)21 compared to (B) The predicted pose for 1 in the comparative model of TbrPDEB1; (C) The TbrPDEB1 homology model (pink) superimposed with hPDE4 (gray). Non-conserved binding site residues are shown as sticks (residues of TbrPDEB1 colored magenta). Human PDE4 numbering is as reported by Card et al.21 (D) View of the P-pocket in the TbrPDEB1 homology model structure, viewed from the opposite face.
Comparison of the active sites in the homology model structures for the two TbrPDEBs confirms that their pairwise conservation is high (see Supporting Information), and that they possess regions previously observed in other PDEs, including a metal binding pocket (M pocket); a solvent-filled side pocket (S pocket); and a pocket containing the conserved purine-binding glutamine residue and hydrophobic clamp (Q pocket), with two hydrophobic pockets (Q1 and Q2) on either side.21 Besides confirming the high similarity in the active site region between TbrPDEB1 and B2, homology modeling also indicates that both are likely to have an extended binding site cleft, termed the “parasite-“ or “P-pocket,” that was first observed in LmjPDEB1.20 This extended pocket provides a tunnel from the binding site leading to the exterior of the protein (Figure 3D), consistent with the P-pocket observed in LmjPDEB1. This feature is absent from every human PDE, and may therefore be exploitable for development of selective inhibitors of parasite PDEs.
While TbrPDEB1 and human PDE4B show overall similarity in the active site, comparison of the homology model with human PDE4-inhibitor crystal structures illustrates multiple differences. These differences may explain the lower inhibitory potency we observe in the trypanosome enzymes, compared to those reported against human PDE4B and PDE4D. For example, the cyclopentyl ring of 1 (and that of rolipram (7) and the cyclopropylmethyl of roflumilast (6)) is buried in the lipophilic Q2 pocket in human PDE4, whereas it is predicted to incompletely fill the P-pocket in TbrPDEB1 and to therefore have more energetically unfavorable exposure to solvent. The Q1 pocket, which accepts the methoxy groups of 1 and 7, and the difluoromethyl group of 6, shows subtle differences in polarity. There are a total of five residues in the Q pocket that are not conserved between human PDE4B and TbrPDE1 that represent significant differences in shape, polarity, hydrogen-bonding capability, hydrophobicity/hydrophilicity, and/or polarizability: P396, Y403, T407, M411, and S442 in human PDE4B are substituted by V826, S833, A837, T841, and G873, respectively, in TbrPDE1 (Figure 3C). These changes are likely to affect the binding properties of the Q-pocket region. Finally, the metal binding pocket is slightly more closed in the TbrPDEB1 model compared to hPDE4B and the observed amino acid difference (Ser786 to Ala348) may alter the interactions between the metal (via intervening water molecules) and the dichloropyridine nitrogen atom of roflumilast and piclamilast or the pyrrolidone headgroup of rolipram. Cumulatively, these relatively small changes may account for the observed differences in potencies between these similar compounds.
In light of the tight structure-activity relationships between piclamilast and its structural congeners roflumilast and rolipram, we wished to further evaluate the impact of minor changes to the piclamilast chemotype. This would allow us to understand the suitability of this series for further optimization. The PDE4B-1 crystal structure21 reveals a pivotal bidentate hydrogen bonding interaction between the invariant binding site glutamine and the catechol ether moiety of the compound; indeed, this binding feature is conserved across all catechol ether-containing PDE inhibitors.21.
In human PDE4B, the cyclopentyl sidechain of 1 is oriented into a small lipophilic pocket in the human enzyme.21 The corresponding pocket in the parasite enzyme, observed in the reported LmjPDEB1 crystal structure20 and also apparent in our homology models, is the deeper “P-pocket” that could potentially accept a larger side-chain from an inhibitor. With this in mind, a focused homologous series of compounds (Entries 2-7) was synthesised to explore this region. Gradual extension of the C-3 alkyl ether moieties to longer chain lengths maintained a level of potency similar to that of 1, yet none were superior. Notably, increase of steric bulk at this position has been reported to reduce hPDE4 potency,22 suggesting that this region of the molecule could be used to improve parasite/host enzyme selectivity. Activity was lost upon further alteration of the catechol substituents (Entries 8-9) and aromatic ring substituents (Entries 10-12). As predicted, there was reasonable agreement between the potency of these inhibitors against TbrPDEB1 and TbrPDEB2.
We advanced a subset of these piclamilast analogs into cellular assays to assess their effects as trypanocides, revealing EC50 values that were in the micromolar range and similar to the IC50 values for TbrPDEB1 and TbrPDEB2 (Table 2, Figure 2). We also selected a compound without TbrPDEB1 or B2 activity, NEU226 (9), to test as a negative control; this compound showed little activity in the cell assay (~10% toxicity at 100 μM).
Table 2.
SAR studies of piclamilast analogs.
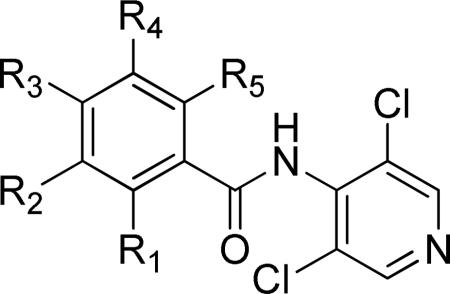 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
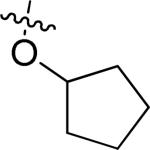
| Entry | Cmpd | R1 | R2 | R3 | R4 | R5 | TbrPDEB1 IC50 (μM)a | TbrPDEB2 IC50 (μM)a | T. brucei EC50 (μM) |
| 1 | 1 | H |
|
OMe | H | H | 4.7 ± 1.0 | 11.4± 1.1 | 9.6 ± 0.9 |
| 2 | 10 22 | H | OMe | OMe | H | H | >100 | >100 | |
| 3 | 11 | H | OEt | OMe | H | H | 16.5±3.4 | 34.0±0.9 | |
| 4 | 12 | H | OPr | OMe | H | H | 13.6± 4.4 | 9.4±2.4 | 13.0±2.4 |
| 5 | 13 | H | OiPr | OMe | H | H | >30b | >30b | 27.6±7.1 |
| 6 | 14 22 | H | OBu | OMe | H | H | 7.7 ± 3.9 | 14.3±3.7 | 17.7±3.1 |
| 7 | 15 | H | OBn | OMe | H | H | 12.5±5.3 | 11.2±1.1 | 10.1±1.2 |
| 8 | 16 | H | OEt | OEt | H | H | >100 | nd | |
| 9 | 17 | H | OMe | OAc | H | H | >100 | nd | |
| 10 | 9 | H | OMe | OMe | OMe | H | >>100 | nd | >>100 |
| 11 | 18 | Cl | OMe | OMe | H | H | >>100 | nd | |
| 12 | 19 | H | OMe | OMe | H | Cl | >>100 | nd | |
IC50 values reported to be “>> x μM” if <25% inh at x μM; “> x μM” if 25-50% inh at x μM.
Measurement of a precise IC50 value was precluded by low compound solubility.
Discussion and conclusions
The PDEB family of trypanosome enzymes has been demonstrated to be essential for parasite growth and virulence9 and as such may provide a route to the development of anti-trypanosomal therapeutics.10 Inhibition of human PDEs has proved to be fertile ground for development of new therapeutics. Previous work targeting human PDEs has established numerous inhibitor series, plus PDE assay methodologies, structural biology information, and, perhaps most importantly, precedence that drug-like PDE inhibitors can be invented. With that in mind, we primarily took the approach of repurposing PDE inhibitors that have at least entered Phase I clinical trials. Benchmarking of inhibitors against human PDEs established several PDE4 compounds as the most potent inhibitors of both TbrPDEB1 and TbrPDEB2. Of these compounds, piclamilast (1) was selected as a starting point for initial medicinal chemistry explorations. In light of the inactivity of the close piclamilast analogs rolipram and roflumilast, we were acutely interested in exploring initial SAR around 1 to ensure that its potency was not an aberration among other PDE4 inhibitors. The identification of piclamilast analogs that retain biochemical and cellular activity provides increased confidence that this series can be pursued for further optimization. This optimism is confirmed by a recent report of a similar catechol ether chemotype as a TbrPDEB1 and LmjPDEB1 inhibitor.23
The most potent compounds identified in the biochemical assay were moved forward into assays of trypanocidal activity. These inhibitors of enzyme activity were cytotoxic in a dose dependent manner, producing micromolar EC50 values, similar to the IC50 values determined against the recombinant TbrPDEBs. This provided confidence that the cell permeability of 1 was maintained in the analogs., Also, that the TbrPDE-inactive compound 9 did not show cellular activity suggests that the trypanocidal effect is likely due to PDE inhibition.
In summary, we report the profiling of a range of human PDE inhibitors against trypanosomal PDEs and describe the activity of a range of analogs of the human PDE4 inhibitor piclamilast (1). This compound and some of its analogs are trypanocidal in a dose-responsive manner. In the context of a target repurposing approach, since the prototypical PDE4 inhibitor rolipram (7) was found to be inactive in our hands and by others, we also show the importance of testing as many known human inhibitor compounds against the pathogen homologs as feasible. In doing so, we have uncovered a potentially optimizable chemotype for inhibition of trypanosomal PDEs. Work is ongoing to improve the potency of these inhibitors and to understand the structural basis of enzyme binding
Experimental Section
Compound procurement
EHNA, milrinone, trequinsin, etazolate, rolipram, Ro 20-1724, dipyridamole, and zaprinast were obtained from Fisher Scientific. Roflumilast was obtained from Carbomer, Inc. BAY 19-8004 was obtained from Axon Medchem, and IBMX was obtained from Sigma-Aldrich. Sildenafil was generously provided by Pfizer, Inc. and was also prepared as previously reported.24 All other compounds were prepared according to their published reports (see Supporting Information for compound structures and reference information).
Vector Construction
DNA synthesis was employed to create open reading frames encoding the catalytic domains of T. brucei brucei PDEB1 (Tb09.160.3590, Ser580-Arg930) and PDEB2 (Tb09.160.3630, Ser580-Ser925) fused to an N-terminal tandem affinity tag composed of 8X polyhistidine, human Propionyl-CoA Carboxylase (Ser635-Leu702) and a TEV protease cleavage site. The ORFs were codon-optimized for expression in Spodoptera frugiperda Sf21 cells, cloned into pFastBac1 using 5' EcoR1 and 3' XhoI, and sequenced to confirm the integrity of the DNA. Recombinant baculovirus stocks were generated using the Bac-to-Bac Baculovirus Expression System (Invitrogen) and expression of proteins with the predicted size were identified by Western blot with anti-His antibody.
Protein expression and purification
Protein was expressed in the Sf21 cell line (Invitrogen). The cells were harvested by centrifugation, lysed by sonication in buffer A (500 mM NaCl, 5 mM Imidazole, 20 mM Tris, 1.25 mM Brij 35, 10 mM Triton X-100, 5 mM Tween 20, pH 7.5), and the lysate was clarified by centrifugation at 40,000 g for 1 hour. The clarified lysate was loaded onto Ni-Sepharose resin (GE Healthcare) at 4°C and washed with twenty column volumes (CV) of buffer A followed by 20 CV buffer A containing 50 mM imidazole. Bound protein was eluted stepwise in Buffer A containing 100 mM, 200 mM, 300 mM, and 500 mM imidazole. Fractions containing enriched protein were pooled, dialyzed against buffer B (20 mM Tris, pH 7.4, 150 mM NaCl), adjusted to 20% glycerol, and snap-frozen in liquid nitrogen prior to storage at -80°C. The proteins were estimated to be 90% pure by SDS-PAGE.
TbrPDE assay conditions
Recombinant TbrPDEB1 (0.25Ig) and TbrPDEB2 (0.5Ig) were assayed at 37°C in 10mM Tris pH 7.4, bovine serum albumin (0.2 mg/ml), 10mM MgCl2, 25IM cAMP (Enzo Lifesciences) and an excess of 5` nucleotidase (>1000U), as determined by titration (Enzo Lifesciences). Reactions were terminated by the addition of BioMol green (Enzo Lifesciences) which was also used to detect changes in the level of phosphate. This was measured by absorbance at 620nm using a Tecan Sunrise plate reader. Inhibitors were dissolved in DMSO and preincubated with the assay mixture at a final DMSO concentration of 2%, 5% or 10% (v/v) for 10 minutes prior to the addition of substrate. All IC50 values were determined at a final concentration of 10% DMSO. Inhibition was determined by the change in the initial velocity relative to a vehicle only control. The KM values were determined as described above with the exception that substrate concentrations varied in the range of 7.8 to 250 IM. The specific activity was determined at 2%, 5% and 10% DMSO by comparison with standards. Standard curves were generated by the reaction of 5` nucleotidase with 0.05 to 3 nmol AMP (Enzo Lifesciences), under conditions identical to those described above, with the exception that PDE was omitted from the reaction. All values are the mean of three or more independent experiments. Data were analyzed using GraphPad Prism 5.0.
Trypanosome cell culture assays
Bloodstream forms of Trypanosoma brucei brucei strain 427 were grown at 37 °C in a 5% CO2 atmosphere in HMI-11 medium25 supplemented with 10% fetal bovine serum (FBS, Sigma). Cells in the mid-logarithmic stage of growth were diluted to a density of 104 cells/ml and were incubated with a range of concentrations of inhibitor in DMSO or DMSO alone. The final concentration of DMSO was 1%. Cell densities were determined after 48 h using Alamar blue (Invitrogen) per the manufacturer's instructions. All values are the mean of three or more independent experiments.
Homology modeling
The protein sequences of TbrPDEB1 and TbrPDEB2 (see Supporting Information) were obtained from TriTrypDB (http://tritrypdb.org/) using the gene identification numbers TbrPDEB1 (Tb09.160.3590) and TbrPDEB2 (Tb09.160.3630). The protein sequences of both proteins were searched against the PDB database (http://www.pdb.org/) using PSI-BLAST.26 Only the catalytic domains of these proteins were used for comparative modeling using the homology feature in the YASARA suite of programs.27 Multiple TbrPDEB1 and TbrPDEB2 models were built using human PDE4 and/or LmjPDEB120 as templates. The best ranked models for both TbrPDEB1 and TbrPDB2 were based on LmjPDEB1 as the template. Multiple modeling evaluation tools were used to confirm the quality of the model structures, including PROCHECK,28 MolProbity,29 and Verify3D,30 described in detail in the Supporting Information.
Piclamilast docking
The model TbPDEB1 structures were further processed using the Maestro 9.1 protein preparation wizard (Schrodinger, LLC, 2010, New York, NY). A restrained minimization of the protein structure was performed using the default constraint of 0.3 Å RMSD and OPLS 2001 force field. The 3D coordinates of piclamilast (1) were then generated using the ligprep utility in Maestro 9.0. The docking parameters were first examined by replication of the crystal structures of the PDE4D/roflumilast complex (PDB ID 1XOQ) and the PDE4B/rolipram complex (PDB ID 1XMY).21 Docking was performed with Glide version 3.5 in standard precision (SP) mode. The docking experiments were conducted with the constraint that at least one H-bond must be formed between the ligand and conserved Gln in the P clamp.
Chemical synthesis
Unless otherwise noted, reagents were obtained from Sigma-Aldrich, Inc. (St. Louis, MO), and used as received. Reaction solvents were purified by passage through alumina columns on a purification system manufactured by Innovative Technology (Newburyport, MA). NMR spectra were obtained Varian NMR systems, operating at 400 or 500 MHz for 1H acquisitions as noted. LCMS analysis was performed using a Waters Alliance reverse-phase HPLC, with single-wavelength UV-visible detector and LCT Premier time-of-flight mass spectrometer (electrospray ionization). All newly synthesized compounds were deemed >95% pure by LCMS analysis prior to submission for biological testing.
General procedure
Various substituted benzoic acids (0.2 mmol) were obtained in pre-weighed quantities in 8 mL screw cap vials. To the acid (1 equiv.) was added 3 mL of thionyl chloride and the mixture was agitated on a heated shaker plate at 90 °C for 3 hours. The crude mixture was concentrated using a Genevac evaporator, and residual thionyl chloride was azeotropically removed with 2-3 sequential additions and evaporations of toluene. In a separate, flame-dried flask, 3,5-dichloropyridin-4-amine (0.75 equiv.) was dissolved in dry THF and then added dropwise to sodium hydride (2 equiv.) in dry THF (0.1 M final concentration) under an inert atmosphere at 0 °C. The mixture was allowed to warm to room temperature and was stirred for 2 hours and cooled once again to 0 °C. The acid chloride prepared above was dissolved in THF (0.1 M final concentration) and added dropwise to the aminopyridine suspension, and the reaction mixture was stirred for 24 hours at room temperature. The solvent was removed by evaporation, and the residue was taken up in EtOAc. The organic layer was washed with 1 M HCl (1x), then NaHCO3 (3x) and dried over Na2SO4. The desired products were isolated following purification via silica gel chromatography (EtOAc/hexanes gradient).
N-(3,5-dichloropyridin-4-yl)-3,4,5-trimethoxybenzamide (9). Yield: 54%. 1H NMR (400 MHz, CDCl3) δ 8.60 (s, 2H), 7.78 (s, 1H), 7.22 (s, 2H), 3.08 (s, 9H). LCMS found 357.01 [M+H]+.
N-(3,5-dichloropyridin-4-yl)-3-ethoxy-4-methoxybenzamide (11). Yield: 12%. 1H NMR (500 MHz, CDCl3) δ 8.56 (s, 2H), 7.64 (br, 1H), 7.52 (s, 1H), 7.5 (m, 1H), 6.95 (d, J = 8.0 Hz, 1H), 4.21 (q, 2H), 3.96 (s, 3H), 1.50 (t, J = 7.0 Hz, 3H). LCMS found 341.01 [M+H]+.
N-(3,5-dichloropyridin-4-yl)-4-methoxy-3-propoxybenzamide (12). Yield: 25%. 1H NMR (400 MHz, d6-DMSO) δ 10.41 (s, 1H), 8.73 (s, 2H), 7.65 (d, J = 8.8 Hz, 1H), 7.54 (d, J = 2.0 Hz, 1H), 7.10 (d, J = 8.8 Hz, 1H), 3.97 (t, J = 6.6 Hz, 2H), 3.84 (s, 3H), 1.75 (m, 2H), 0.97 (t, J = 7.6 Hz, 3H). . LCMS found 355.01 [M+H]+.
N-(3,5-dichloropyridin-4-yl)-3-isopropoxy-4-methoxybenzamide (13). Yield: 37%. 1H NMR (400 MHz, CDCl3) δ 8.54 (s, 2H), 7.71 (s, 1H), 7.53 (m, 1H), 7.50 (d, J = 2.0 Hz, 1H), 6.94 (d, J = 8.8 Hz, 1H), 4.64 (m, 1H), 3.93 (s, 1H), 1.40 (d, J = 6.0 Hz, 1H). LCMS found 355.01 [M+H]+.
3-(benzyloxy)-N-(3,5-dichloropyridin-4-yl)-4-methoxybenzamide (15). Yield: 22%. 1H NMR (400 MHz, CDCl3) δ 8.54 (s, 2H), 7.59 (s, 1H), 7.53 (m, 2H), 7.46 (d, J = 7.2 Hz, 2H), 7.37 (t, J = 7.2 Hz, 2H), 7.32 (d, J = 7.2 Hz, 1H), 6.97 (d, J = 8 Hz, 1H), 5.21 (s, 2H), 3.95 (s, 3H). LCMS found 403.01 [M+H]+.
N-(3,5-dichloropyridin-4-yl)-3,4-diethoxybenzamide (16). Yield: 50%. 1H NMR (400 MHz, CDCl3) δ 8.54 (s, 2H), 7.71 (s, 1H), 7.51-7.47 (m, 2H), 6.92 (d, J = 8.0 Hz, 1H), 4.16 (m, 4H), 1.48 (t, 6H). LCMS found 355.01 [M+H]+.
4-((3,5-dichloropyridin-4-yl)carbamoyl)-2-methoxyphenyl acetate (17). Yield: 45%. 1H NMR (400 MHz, CDCl3) δ 8.57 (s, 2H), 7.71 (s, 1H), 7.60 (m, 1H), 7.50 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.0 Hz, 1H), 3.93 (s, 3H), 2.35 (s, 3H). LCMS found 355.01 [M+H]+.
2-chloro-N-(3,5-dichloropyridin-4-yl)-3,4-dimethoxybenzamide (18). Yield: 49%. 1H NMR (400 MHz, CDCl3) δ 8.55 (s, 2H), 8.25 (s, 1H), 7.71 (d, J = 8.8 Hz, 1H), 6.93 (d, J = 8.8 Hz, 1H), 3.94 (s, 3H), 3.89 (s, 3H). LCMS found 361.01 [M+H]+.
2-chloro-N-(3,5-dichloropyridin-4-yl)-4,5-dimethoxybenzamide (19). Yield: 51%. 1H NMR (500 MHz, d6-DMSO) δ 10.65 (s, 1H), 8.73 (s, 2H), 7.41 (s, 1H), 7.13 (d, J = 7.5 Hz, 1H), 3.82 (s, 3H), 3.80 (s, 3H). LCMS found 361.01 [M+H]+.
Supplementary Material
Acknowledgements
Funding from the National Institutes of Health (R01AI082577, including an ARRA summer supplement), the National Science Foundation (MCB-0843603), Northeastern University, Boston University, and gift of sildenafil from Pfizer, Inc. are gratefully acknowledged.
Abbreviations
- HAT
human African trypanosomiasis
- BF
bloodstream form T. brucei
- cAMP
cyclic adenosine monophosphate
- cGMP
cyclic guanosine monophosphate
- PDB
Protein Data Bank
- PDE
Phosphodiesterase
- TbrPDEB1
PDEB1 of Trypanosoma brucei
- TbrPDEB2
PDEB2 of Trypanosoma brucei
- CNS
central nervous system
- NTD
neglected tropical disease
- EHNA
erythro-9-(2-hydroxy-3-nonly)adenine
Footnotes
Supporting Information. A tabulation of all the benchmarked human PDE inhibitors, with their references and screening data is in the Supporting Information, and is publically available as a shared data set at www.collaborativedrugdiscovery.com. The Supporting Information also includes protein sequences of TbrPDEB1 and B2, and alignment between TbrPDEB1, B2, LmjPDEB1, and hPDE4D. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N. Engl. J. Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 2.Croft SL, Barrett MP, Urbina JA. Chemotherapy of trypanosomiases and leishmaniasis. Trends in Parasitology. 2005;21:508–512. doi: 10.1016/j.pt.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 3.Pollastri MP, Campbell RK. Target repurposing for neglected diseases. Future Med. Chem. 2011;3:1307–1315. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Kumar G, Kumar RS. An update on cyclic nucleotide phosphodiesterase (PDE) inhibitors: phosphodiesterases and drug selectivity. Methods Find. Exp. Clin. Pharmacol. 2005;27:101–118. doi: 10.1358/mf.2005.27.2.876285. [DOI] [PubMed] [Google Scholar]
- 5.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: A new target for the development of specific therapeutic agents. Pharmacol. Therap. 2006;109:366–398. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Martin H, Christopher K. Prof. Dr. Hugo Kubinyi DGM, editor. Chemogenomics in Drug Discovery. 2005:243–288. [Google Scholar]
- 7.Menniti FS, Faraci WS, Schmidt CJ. Phosphodiesterases in the CNS: targets for drug development. Nat. Rev. Drug. Discov. 2006;5:660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 8.Gould MK, de Koning HP. Cyclic-nucleotide signalling in protozoa. FEMS Microbiol. Rev. 2011 doi: 10.1111/j.1574-6976.2010.00262.x. [DOI] [PubMed] [Google Scholar]
- 9.Oberholzer M, Marti G, Baresic M, Kunz S, Hemphill A, Seebeck T. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 2007;21:720–731. doi: 10.1096/fj.06-6818com. [DOI] [PubMed] [Google Scholar]
- 10.Shakur Y, de Koning HP, Ke H, Kambayashi J, Seebeck T. Therapeutic potential of phosphodiesterase inhibitors in parasitic diseases. Handb. Exp. Pharmacol. 2011:487–510. doi: 10.1007/978-3-642-17969-3_20. [DOI] [PubMed] [Google Scholar]
- 11.Rascon A, Soderling SH, Schaefer JB, Beavo JA. Cloning and characterization of a cAMP-specific phosphodiesterase (TbPDE2B) from Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2002;99:4714–4719. doi: 10.1073/pnas.002031599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoraghi R, Seebeck T. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2002;99:4343–4348. doi: 10.1073/pnas.062716599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beghyn TB, Charton J, Leroux F, Laconde G, Bourin A, Cos P, Maes L, Deprez B. Drug to Genome to Drug: Discovery of New Antiplasmodial Compounds. J. Med. Chem. 2011 doi: 10.1021/jm1014617. [DOI] [PubMed] [Google Scholar]
- 14.Laxman S, Rascon A, Beavo JA. Trypanosome cyclic nucleotide phosphodiesterase 2B binds cAMP through its GAF-A domain. J. Biol. Chem. 2005;280:3771–3779. doi: 10.1074/jbc.M408111200. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chappie TA, Humphrey JM, Allen MP, Estep KG, Fox CB, Lebel LA, Liras S, Marr ES, Menniti FS, Pandit J, Schmidt CJ, Tu M, Williams RD, Yang FV. Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors. J. Med. Chem. 2007;50:182–185. doi: 10.1021/jm060653b. [DOI] [PubMed] [Google Scholar]
- 17.Dyke HJ, Montana JG. Update on the therapeutic potential of PDE4 inhibitors. Expert Opin. Invest. Drugs. 2002;11:1–13. doi: 10.1517/13543784.11.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Montana JG, Dyke HJ. Phosphodiesterase 4 inhibitors. Annu. Rep. Med. Chem. 2001;36:41–56. [Google Scholar]
- 19.Pages L, Gavalda A, Lehner MD. PDE4 inhibitors: a review of current developments (2005 - 2009). Expert Opin. Ther. Pat. 2009;19:1501–1519. doi: 10.1517/13543770903313753. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Yan Z, Geng J, Kunz S, Seebeck T, Ke H. Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors. Mol. Microbiol. 2007;66:1029–1038. doi: 10.1111/j.1365-2958.2007.05976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Card GL, England BP, Suzuki Y, Fong D, Powell B, Lee B, Luu C, Tabrizizad M, Gillette S, Ibrahim PN, Artis DR, Bollag G, Milburn MV, Kim S-H, Schlessinger J, Zhang KYJ. Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases. Structure. 2004;12:2233–2247. doi: 10.1016/j.str.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Ashton MJ, Cook DC, Fenton G, Karlsson J-A, Palfreyman MN, Raeburn D, Ratcliffe AJ, Souness JE, Thurairatnam S, Vicker N. Selective Type IV Phosphodiesterase Inhibitors as Antiasthmatic Agents. The Syntheses and Biological Activities of 3-(Cyclopentyloxy)-4-methoxybenzamides and Analogs. J. Med. Chem. 1994;37:1696–1703. doi: 10.1021/jm00037a021. [DOI] [PubMed] [Google Scholar]
- 23.Seebeck T, Sterk GJ, Ke H. Phosphodiesterase inhibitors as a new generation of antiprotozoan drugs: exploiting the benefit of enzymes that are highly conserved between host and parasite. Future Med. Chem. 2011;3:1289–1306. doi: 10.4155/fmc.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terrett NK, Bell AS, Brown D, Ellis P. Sildenafil (VIAGRA), a potent and selective inhibitor of type 5 cGMP phosphodiesterase with utility for the treatment of male erectile dysfunction. Bioorg. Med. Chem. Lett. 1996;6:1819–1824. [Google Scholar]
- 25.Hirumi H, Martin S, Hirumi K, Inoue N, Kanbara H, Saito A, Suzuki N. Cultivation of bloodstream forms of Trypanosoma brucei and T. evansi in a serum-free medium. Trop. Med. & Int. Health. 1997;2:240–244. doi: 10.1046/j.1365-3156.1997.d01-268.x. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK - A program to check the stereochemical quality of protein structures. J. Appl. Crystallog. 1993;26:283–291. [Google Scholar]
- 29.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: Assessment of protein models with three-dimensional profiles. Macromol. Crystallog. Pt. B. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 31.Braeunlich G, Fischer R, Es-sayed M, Henning R, Sperzel M, Schlemmer K-H, Nielsch U, Tudhope S, Sturton G. Preparation of N-(3-benzofuranyl)ureas as antiinflammatory agents. 1996 et, a. EP731099A1.
- 32.Gruenman V, Hoffer M. 4-Benzyl-2-imidazolidinones from N-[(1-cyano-2-phenyl)ethyl] carbamates. 1975 US3923833A.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.