Figure 4.
TFL2/LHP1 Acts in Floral Stem Cell Termination, and WUS Is a PcG Target.
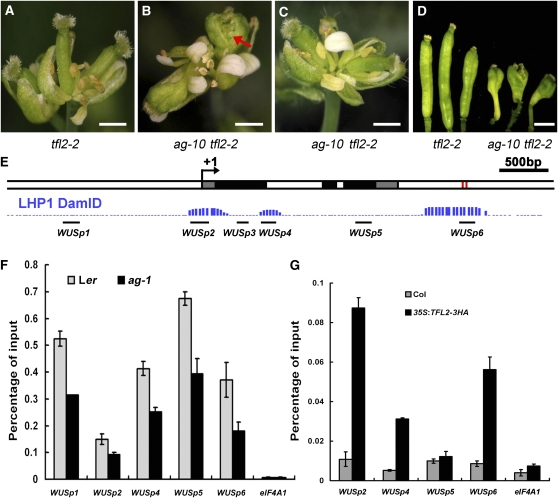
(A) A terminal inflorescence composed of several fused flowers in tfl2-2. Note that the gynoecia were thin.
(B) A representative inflorescence of ag-10 tfl2-2 plants with floral determinacy defects. The flowers had bulged gynoecia with ectopic floral organs inside (arrow).
(C) A representative inflorescence of ag-10 tfl2-2 plants without floral determinacy defects. The gynoecia were thin.
(D) Siliques from plants of the indicated genotypes. The ag-10 tfl2-2 plants were from the F2 population of the cross between ag-10 and tfl2-2. Only siliques from the ag-10 tfl2-2 plants with floral determinacy defects are shown. Bars = 1 mm in (A) to (D).
(E) A diagram of the WUS genomic region with “+1” being the transcription start site. Gray, black, and white rectangles represent 5′ or 3′ untranslated regions, coding regions, and introns or intergenic regions, respectively. The two red rectangles represent the two CArG boxes. The three regions of TFL2/LHP1 occupancy at WUS as determined by genome-wide profiling of TFL2/LHP1 binding sites are shown in blue (LHP1 DmID; Zhang et al., 2007a). The regions interrogated for AG, H3K27me3, or TFL2/LHP1 enrichment at WUS in this study are shown as black bars.
(F) ChIP with anti-H3K27me3 antibodies to determine the levels of H3K27me3 at WUS in wild-type (Ler) and ag-1 inflorescences containing stage 8 and younger flowers.
(G) ChIP with anti-HA antibodies in Col (a negative control) and 35S:TFL2-3HA to examine TFL2/LHP1 occupancy at WUS. For (F) and (G), the regions examined are diagramed in (E). eIF4A1 served as a negative control. Error bars represent sd, which were calculated from three technical repeats. Three biological replicates gave similar results.