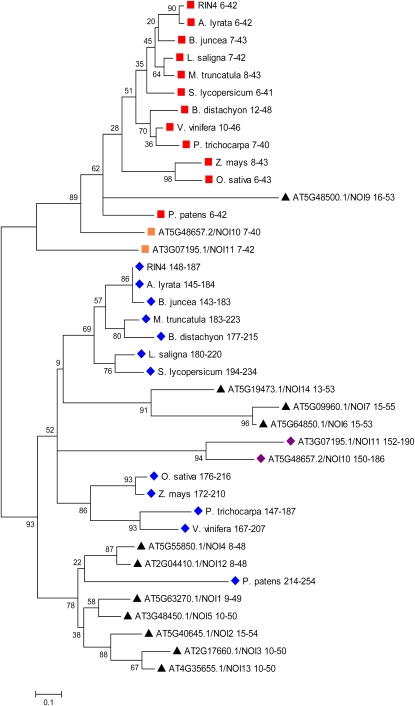
Figure 3.
The N-NOI and C-NOI of RIN4 Are from Evolutionarily Distinct Clades.
The NOI domains from RIN4, its closest BLAST homolog from a variety of plants, and other NOI-containing proteins from Arabidopsis (NOI1 to NOI14) were analyzed. The NOI domains were aligned by ClustalW, and their evolutionary relationship was inferred by the Neighbor-Joining method using MEGA4. The confidence probability (×100) that the interior branch length is greater than 0, as estimated using the bootstrap test (1000 replicates), is shown next to the branches. Branch lengths represent evolutionary distances. Number ranges are the amino acid positions of each NOI within its respective protein. The N-NOIs and C-NOIs from RIN4 homologs from different plant species are indicated by red squares and blue diamonds, respectively. Orange squares and purple diamonds indicate the N-NOIs and C-NOIs from Arabidopsis NOI proteins with a structure similar to RIN4 (NOI10 and NOI11). Black triangles indicate the NOIs from Arabidopsis proteins with only a single NOI domain. All NOIs start with an AvrRpt2 cleavage site, with the exception of the C-NOIs of NOI10 and NOI11 from Arabidopsis, which have a Pro insertion and Trp substitution predicted to disrupt cleavage by AvrRpt2. NOI8 (At5G18310) is excluded from the analysis, because ambiguous gene models exist in TAIR9. RIN4 is At3G25070. For other Arabidopsis proteins containing NOIs, the AGI numbers are shown. RIN4 homologs are from the following plant species: A. lyrata, B. juncea, M. truncatula, L. saligna, S. lycopersicum, P. trichocarpa, V. vinifera, O. sativa, Z. mays, B. distachyon, and P. patens.