Figure 3.
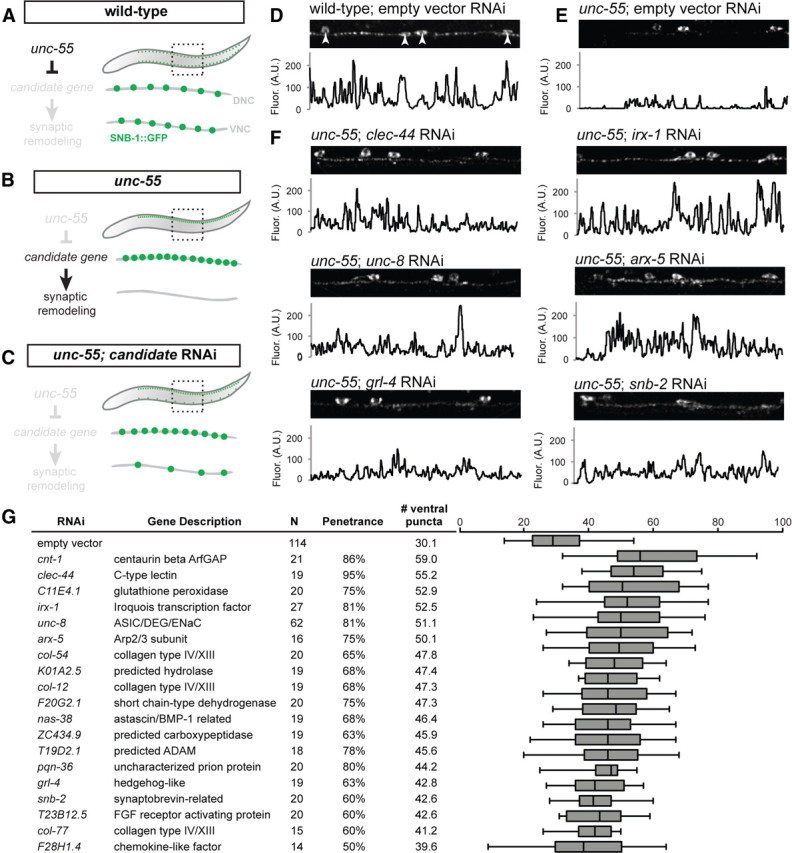
RNAi of candidate UNC-55 targets identifies genes required for synaptic remodeling. A–C, Experimental design of Unc-55 suppression screen. A, In wild-type animals, SNB-1::GFP puncta (marking GABAergic NMJs) localize to both dorsal and ventral nerve cords. B, In unc-55 animals, synaptic remodeling genes are de-repressed in VD motor neurons and SNB-1::GFP is depleted from the ventral nerve cord. C, RNAi knockdown of UNC-55 target genes in unc-55 mutants disrupts synaptic remodeling and restores SNB-1::GFP puncta to the ventral nerve cord. D–F, Representative views (50 μm length) of GABAergic SNB-1::GFP in adult ventral nerve cords. Line scans (in arbitrary fluorescence units) of the ventral nerve cord (excluding DD/VD cell bodies; D, arrowheads). D, SNB-1::GFP puncta are abundant in the ventral nerve cord of wild-type adults (E) but largely absent from unc-55(e1170) mutant animals treated with control RNAi (empty vector). F, RNAi knockdown of candidate unc-55 targets (clec-44, irx-1, unc-8, arx-5, grl-4, snb-2) in unc-55 mutants partially restores ventral SNB-1::GFP puncta. G, Conserved hits from Unc-55 RNAi suppression screen. Hits were scored as significantly different from empty vector control (p < 0.01, Student's t test) with restoration of ventral SNB-1::GFP puncta in ≥50% of animals scored. N represents all animals scored, combined from at least two separate RNAi treatments (see Materials and Methods). The boxes on the box-and-whisker plot span the 25th to 75th percentile, and the whiskers indicate the minimum/maximum number of ventral puncta observed; the vertical line in each box indicates the 50th percentile. Conserved hits show <e−10 versus human homologs by BLAST.
