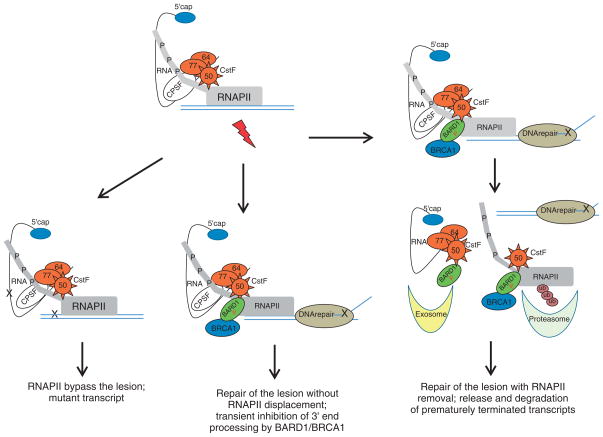
FIGURE 2.
Model for transcriptional–3′-end processing alternatives during the DNA damage response (DDR). The DNA damage-induced lesions may affect the elongating RNA polymerase II–cleavage stimulation factor (RNAP II–CstF) holoenzyme in different ways. The lesion is bypassed, generating a mutant transcript. Alternatively, RNAP II stalled at sites of DNA damage can reengage in a transcriptional process. In that scenario, the BRCA1–BARD1–CstF complex reassures the correct 3′-end processing of the nascent RNA. For some other lesions, RNAP II–CstF holoenzyme is stalled, causing premature termination or transient arrest of elongation process. BRCA1–BARD1-containing complexes are recruited to sites of DNA repair, resulting in RNAP II ubiquitination and inhibition of transcription by degradation of RNAP II. The nascent RNA product is released and its 3′-end processing is inhibited by both the CstF–BARD1 interaction and RNAP II degradation. This process facilitates repair by allowing access of the repair machinery to the DNA damage site, and at the same time prevents the formation of aberrantly processed mRNAs, which are eliminated by exosome-mediated degradation in a nuclear surveillance pathway.