Abstract
Plant development and productivity are negatively regulated by adverse environmental conditions. The identification of stress-regulatory genes, networks, and signaling molecules should allow the development of novel strategies to obtain tolerant plants. Polyamines (PAs) are polycationic compounds with a recognized role in plant growth and development, as well as in abiotic and biotic stress responses. During the last years, knowledge on PA functions has been achieved using genetically modified plants with altered PA levels. In this review, we combine the information obtained from global transcriptome analyses in transgenic Arabidopsis plants with altered putrescine or spermine levels. Comparison of common and specific gene networks affected by elevation of endogenous PAs, support the view that these compounds actively participate in stress signaling through intricate crosstalks with abscisic acid (ABA), Ca2+ signaling and other hormonal pathways in plant defense and development.
Involvement of Polyamines in Plant Responses to Environmental Stimuli
Environmental conditions that promote plant stress adversely affect their growth and productivity by triggering a series of morphological, physiological, biochemical, and molecular changes. Use of modern molecular biology tools for elucidating the control mechanisms of abiotic stress tolerance and development of strategies to obtain stress-tolerant plants are currently one of the most active fields in plant research, which helps to prevent the dramatic reduction in crop yields due to global warming effects. Polyamines (PAs) are small protonated compounds with key roles in plant development and stress protection, of which the most predominant forms are the diamine putrescine (Put), triamine spermidine (Spd), and tetramine spermine (Spm). Elevated PA levels have been observed in different plant species in response to salinity, drought, chilling, heat, hypoxia, ozone, UV-B, and UV-C, heavy metal toxicity, mechanical wounding, and herbicide treatment (reviewed in Alcázar et al., 2006b, 2010a; Bouchereau et al. 1999; Groppa and Benavides 2008). Most of the studies conclude that PA accumulation represents a stress-induced response with a protective role. However, the precise mechanism(s) of action by which PAs could protect plants from challenging environmental conditions remains unclear, although some progress has been made (Alcázar et al., 2010a; Gill and Tuteja, 2010).
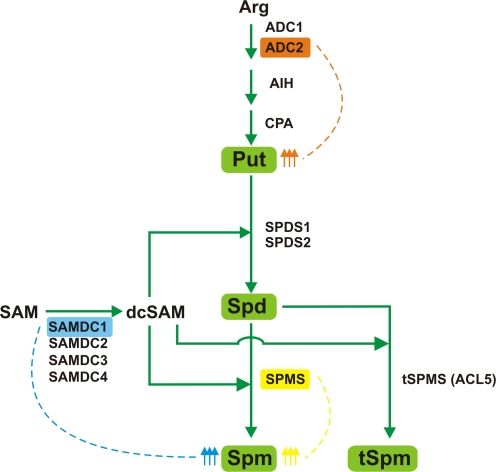
PA biosynthesis pathway has been established in detail in Arabidopsis thaliana (Alcazar et al., 2006b). PA synthesis starts with the synthesis of the diamine Put. This step is done in Arabidopsis by decarboxylation of arginine (Arg) by arginine decarboxylase (ADC), and two additional successive steps involving agmatine iminohydrolase (AIH) and N-carbamoylputrescine amidohydrolase (CPA) activities. Higher molecular weight PAs (Spd and Spm) are formed by sequential additions of aminopropyl groups to Put and Spm, respectively, by the activities of Spd synthase (SPDS) and Spm synthase (SPMS). Both enzymes use decarboxylated S-adenosylmethionine (dcSAM) as donor of aminopropyl moieties, resulting from the decarboxylation of SAM in a reaction catalyzed by SAM decarboxylase (SAMDC). Thermospermine (tSpm), a structural isomer of spermine, is also synthesized from spermidine and dcSAM by tSpm synthase (tSPMS) activity (Fig. 1). The Arabidopsis genome contains two genes encoding ADC (ADC1 and ADC2) (Watson and Malmberg, 1996; Watson et al., 1997) and one for each AIH and CPA (Janowitz et al., 2003; Piotrowski et al., 2003). There are also two genes encoding spermidine synthases, SPDS1 and SPDS2 (Hanzawa et al., 2002), one coding for spermine synthase, SPMS (Panicot et al., 2002), another one coding for thermospermine synthase, ACL5 (Kakehi et al., 2008; Knott et al., 2007), and at least four coding for S-adenosylmethionine decarboxylases, SAMDC1-4 (Urano et al., 2003).
FIG. 1.
Polyamine biosynthesis pathway in Arabidopsis thaliana. Effects of ADC2, SAMDC1, or SPMS overexpression: overexpressed genes on studies cited in this review are highlighted in boxes; arrows indicate the effects on PA levels driven by overexpression of different PA biosynthesis genes. ACL5, ACAULIS5; ADC: arginine decarboxylase; AIH: agmatine iminohydrolase; CPA: N-carbamoylputrescine amidohydrolase; dcSAM: decarboxylated S-adenosylmethionine; SAM: S-adenosylmethionine; SAMDC: S-adenosylmethionine decarboxylase; SPDS: spermidine synthase; SPMS: spermine synthase. tSPMS: thermospermine synthase.
More recently, global “omic” approaches have also been applied to study the function of PAs in response to abiotic stresses. The expression of PA biosynthesis genes is affected by a number of abiotic stresses. Drought induces expression of ADC2, SPDS1, and SPMS (Alcazar et al., 2006a); ADC1, ADC2, and SAMDC2 expression are also induced by cold (Cuevas et al., 2008, 2009; Urano et al., 2003). The manipulation of PA levels by transgenic approaches and the use of loss or gain-of-function mutations has gained knowledge about their roles in plant stress in addition to their antioxidant and structure-stabilizing properties (Alcázar et al., 2010a; Gill and Tuteja, 2010).
In this work, we have studied transcriptome profiles in Arabidopsis transgenic plants with altered PA metabolism in order to identify common and specific gene networks affected by alteration of Put or Spm levels. Our results further reinforce previous evidences showing that PAs are regulatory signaling molecules in intricate crosstalks with hormonal pathways responsive to environmental changes, and that different PAs can trigger a differential transcriptional response.
Transcriptome analysis of Arabidopsis plants with increased Put levels
Transgenic lines with elevated Put Levels were obtained by constitutive homologous overexpression of ADC2 in Arabidopsis (Alcazar et al., 2005). These lines are resistant to freezing (Tiburcio et al., 2009) and drought stress (Alcázar et al. 2010b). Comparison of Affymetrix ATH1 microarray transcriptome profiles of ACD2 overexpressing and wild-type Col-0 plants by Affymetrix Microarray Suite 5.0 statistical algorithms, reveals that 1,608 genes are upregulated and 2,653 downregulated in these lines with elevated Put content (Alcazar et al., 2005). Categorization of both set of genes using the functional enrichment analysis tool GENECODIS 2.0 (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009) shows that the set of overexpressed genes is enriched in those involved in responses to biotic and abiotic stresses as well as auxin-related genes (Table 1).
Table 1.
Functional Enrichment Analysis of Differentially Expressed Genes in Put Overproducer Plants
| GO-term | Description | Induced (1608) | Total (22,604) | p-value Adj | |
|---|---|---|---|---|---|
| Biological Process | |||||
| Defense related | GO:0009651 | response to salt stress | 52 | 306 | 6.67E-07 |
| GO:0046686 | response to cadmium ion | 53 | 318 | 7.12E-07 | |
| GO:0009617 | response to bacterium | 18 | 54 | 1.69E-06 | |
| GO:0006979 | response to oxidative stress | 29 | 143 | 2.12E-05 | |
| GO:0009409 | response to cold | 35 | 202 | 6.06E-05 | |
| GO:0010200 | response to chitin | 25 | 127 | 1.85E-04 | |
| GO:0050832 | defense response to fungus | 15 | 59 | 6.60E-04 | |
| GO:0009408 | response to heat | 20 | 109 | 3.67E-03 | |
| GO:0009612 | response to mechanical stimulus | 5 | 8 | 3.39E-03 | |
| GO:0009611 | response to wounding | 21 | 119 | 3.48E-03 | |
| GO:0051707 | response to other organism | 12 | 55 | 1.31E-02 | |
| GO:0046688 | response to copper ion | 5 | 11 | 1.78E-02 | |
| Hormone related | GO:0009733 | response to auxin stimulus | 36 | 256 | 3.59E-03 |
| GO:0009741 | response to brassinosteroid stimulus | 7 | 17 | 3.25E-03 | |
| GO:0009684 | indoleacetic acid biosynthetic process | 4 | 8 | 3.51E-02 | |
| Other | GO:0006412 | translation | 102 | 365 | 1.04E-31 |
| GO:0042254 | ribosome biogenesis | 32 | 96 | 1.21E-11 | |
| GO:0006605 | protein targeting | 5 | 8 | 3.39E-03 | |
| GO:0015031 | protein transport | 8 | 32 | 3.43E-02 | |
| GO:0006869 | lipid transport | 17 | 99 | 1.69E-02 | |
| GO:0016126 | sterol biosynthetic process | 7 | 23 | 2.22E-02 | |
| GO:0006556 | S-adenosylmethionine biosynthetic process | 3 | 4 | 3.55E-02 | |
| Molecular function | |||||
| GO:0003735 | structural constituent of ribosome | 112 | 364 | 1.49E-39 | |
| GO:0008121 | ubiquinol-cytochrome-c reductase activity | 5 | 8 | 1.70E-02 | |
| GO:0016762 | xyloglucan:xyloglucosyl transferase activity | 5 | 9 | 2.40E-02 | |
| GO-term | Description | Repressed (2653) | Total (22,604) | p-value Adj | |
|---|---|---|---|---|---|
| Molecular function | |||||
| GO:0016987 | sigma factor activity (MF) | 6 | 6 | 1.41E-03 | |
Enrichment analysis of differentially expressed genes in ADC2 overexpressing Arabidopsis plants compared to wild-type Col-0 plants is shown. Analysis was performed with the GENECODIS 2.0 software (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009). (Red: upregulated; green: downregulated. Adj p-value: p-value obtained from hypergeometric distribution test and corrected with false discovery rate method of Benjamini and Hochberg).
Both correlations are also observed when the distribution of differentially expressed genes on ADC2 overexpressing plants is examined with the MAPMAN tool (Thimm et al., 2004; Usadel et al., 2005). The expression of a significant number of hormone and signaling related genes is altered in these lines (Additional data, Supplementary Tables S1 and S2). Among them, genes of the IAA biosynthesis pathway (NIT2, GH3.3, GH3.5, YDK1, IAR3, ILL6), auxin transport (PIN3), and genes coding for up to 20 auxin responsive proteins (Additional data, Supplementary Table S1). Also, genes coding for ethylene biosynthesis enzymes as well as several ethylene-responsive transcription factors are up- and downregulated (Additional data, Supplementary Table S1). In addition, a set of genes coding for some of the ABA biosynthesis enzymes (ABA1, NCDE3, NCDE4) and several ABA responsive genes such as transcription factors ABI1, ABF3, ABF4 appear downregulated, while other ABA putative responsive proteins appeared upregulated (Additional data, Supplementary Table S1). Changes were also observed for biotic stress related hormones. Jasmonate-induced proteins (putative jacalin lectin family proteins) were underexpressed, while two JA pathway genes (OPR2 and OPR3) were overexpressed (Additional data, Supplementary Table S1). Expression changes were also observed for several members of the S-adenosyl-L-methionine:carboxyl-methyltransferase protein family, involved in SA biosynthesis (Additional data, Supplementary Table S1). MAPMAN analysis also reveals that another set of genes with significant changes in expression corresponds to signaling-related genes (Additional data, Supplementary Fig. S1), most of them probably involved in stress responses, as suggested by their E-Northern BAR expression profiles (Additional data, Supplementary Fig. S1) as well as by the overrepresentation of some stress-related biological process annotations when GENECODIS enrichment analysis is applied to these particular gene subsets (data not shown).
Transcriptome analysis of Arabidopsis plants with increased Spm levels
Arabidopsis plants with increased Spm levels have been obtained by transgenic homologous overexpresssion of SAMDC1 gene (Marco et al., in press) or the SPMs gene (González et al., 2011). Elevated Spm levels, produced by homologous overexpression of SAMDC1 in Arabidopsis, enhances tolerance to drought and saline stress (Marco et al., in press). In contrast, Arabidopsis acl5/spms mutant plants, are unable to produce Spm and are hypersensitive to salt and drought stress, while exogenous addition of Spm suppresses these phenotypes (Yamaguchi et al., 2006, 2007). Transcriptome profiles have been obtained by Affymetrix ATH1 microarrays for both SAMDC1 and SPMS overexpressing plants (González et al., 2011; Marco et al., in press). When Significance Analysis of Microarrays software (SAM 3.0) (Tusher et al., 2001) is applied to both transcriptome data, different sets of genes with significant variations with respect to wild-type Col-0 plants are obtained for each type of Spm overproducer: 4,651 (2,083 upregulated and 2,568 downregulated) for 35S-SAMDC1 plants and 3,080 transcripts (1,132 upregulated and 1,948 downregulated) for 35S-SPMs plants (Table 2). GENECODIS functional enrichment analysis applied to both sets of genes shows again that the set of overexpressed genes for both types of Spm-overproducer plants appears enriched in annotation categories involved in defense-related processes from both biotic and abiotic stresses (Additional data, Supplementary Tables S2 and S3). Comparison of both transcriptomes shows that, although both SAMDC1 and SPMS overexpressing plants accumulate Spm, their transcriptome profiles differ. Only 234 upregulated and 333 downregulated genes are in common, which represents 12.2% and 18.4% of genes with significant variations in SAMDC1 and SPMS overexpressing plants, respectively (Table 2). Functional enrichment analysis of the upregulated genes indicates that the most predominant biological processes affected are related to biotic and abiotic stresses, and hormonal pathways in JA biosynthesis and JA/SA responses (Table 2). Moreover, two JA biosynthesis genes are upregulated in both SAMDC1 and SPMS overexpressing plants, as well as NCDE3, a key gene in the ABA biosynthesis pathway (Additional data, Supplementary Table S4). Common signaling-involved genes include 23 receptor-like kinases, three MAP (mitogen-activated protein) kinases and seven genes involved in calcium regulation, all being mainly upregulated. On the other hand, there is also a group of 60 common genes coding for transcription factors (Additional data, Supplementary Fig. S2). Enrichment analysis and BAR E-northern stress profiles of those set of common genes again suggests that Spm overproduction affects stress signaling responses (Additional data, Supplementary Fig. S2). Expression patterns for the majority of common genes are similar in both Spm-overproducer plants compared with wild-type, being the more pronounced differences in the case of SAMDC1 overexpressing lines. In summary, transcriptome analysis shows that elevated Spm levels have an important effect on stress response mechanisms of the plant. This result agrees with the observations done by Mitsuya et al. (2009) where external Spm treatment modulates the expression of a large number of defense-related genes.
Table 2.
Functional Enrichment Analysis of Differentially Expressed Genes in Spm Overproducer Plants
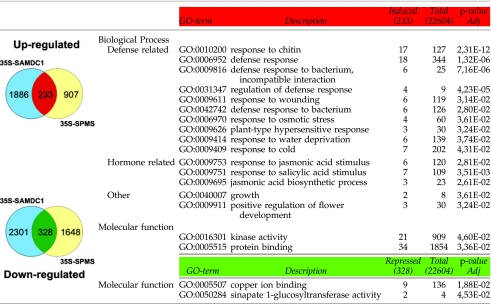 |
Left: venn diagrams showing common genes from differentially expressed genes in SAMDC1 and SPMS overexpressing plants compared to wild-type Col-0 plants. Right: functional enrichment analysis of commonly differentially expressed genes both in SAMDC1 and SPMS overexpressing Arabidopsis plants compared to wild-type Col-0 plants. Analysis was carried out with the GENECODIS 2.0 software (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009). (Red: upregulated; green: downregulated. Adj p-value: p-value obtained from hypergeometric distribution test and corrected with false discovery rate method of Benjamini and Hochberg.)
Transcriptome stress response is altered by modification of PA levels: common responses between Put and Spm overexpressors
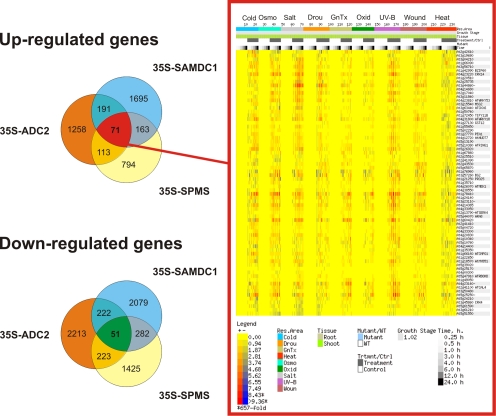
When transcriptome profiles of Put and Spm-overeproducers are compared, only a set of 150 genes with significant expression changes appears in common between the Put and Spm accumulating plants. From this set, 71 genes always appear upregulated, and enriched in stress-related genes (Fig. 2).
FIG. 2.
Expression profiles of common upregulated genes in PA overproducer plants. Left: Venn diagram analysis of common up- and downregulated genes from ADC2, SAMDC1, and SPMS overexpressor plants. Values represent the number of transcripts over and underexpressed in transgenic plants relative to wild-type Col-0 plants. Red box: E-northern heat map analysis of the 71 common upregulated genes in the three PA overproducer plants using abiotic stress series from BAR (Toufighi et al., 2005). Rows represent genes and columns represent experimental conditions. The color at a point represents the log2 of the ratio of the average of replicate treatments relative to the average of corresponding controls. The scale is shown in the figures.
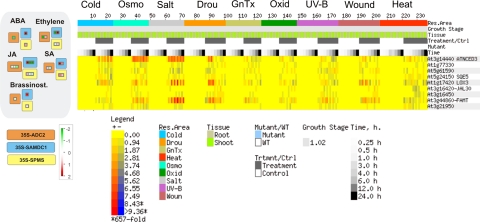
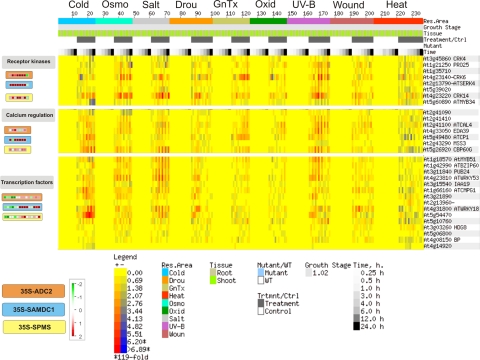
MAPMAN analysis of the expression profiles of these 150 common genes shows similar expression patterns for the majority of common genes represented. On the other hand, the majority of gene sets considered are composed by single or few genes (Fig. 3), except for signaling-related genes, where seven genes involved in calcium signaling have similar expression profiles in Put and Spm overproducer plants (Fig. 4). Among common signaling-related genes, most are also stress-responsive, as shown by E-northern BAR analysis (Fig. 4), as well as by Enrichment analysis (data not shown). The same relationship with stress is shown also for common hormone-related genes (Fig. 3), including NCED3 (Schwartz et al., 1997) and LOX3 (Bannenberg et al., 2009), which code for ABA and JA biosynthesis enzymes, respectively.
FIG. 3.
Comparative analysis of expression profiles of common differentially expressed genes from ADC2, SAMDC1, and SPMS overexpressing plants. Only hormone-related genes are shown. Left: MAPMAN analysis (Thimm et al., 2004; Usadel et al., 2005). The signal from transgenic plants is expressed as a ratio relative to the signal in wild-type Col-0 plants converted to a log2 scale, and displayed. Right: Heatmap expression analysis using abiotic stress series from BAR (Toufighi et al., 2005). Rows represent genes and columns represent experimental conditions. The colour at a point represents the log2 of the ratio of the average of replicate treatments relative to the average of corresponding controls. The scale is shown in the figures. Orange boxes: ADC2 overexpressing plants. Blue boxes: SAMDC1 overexpressing plants; yellow boxes: SPMS overexpressing plants.
FIG. 4.
Comparative analysis of expression profiles of common differentially expressed genes from ADC2, SAMDC1, and SPMS overexpressing plants. Only signaling-related genes are shown. Left: MAPMAN analysis (Thimm et al., 2004; Usadel et al., 2005). The signal from transgenic plants is expressed as a ratio relative to the signal in wild-type Col-0 plants converted to a log2 scale, and displayed. Right: Heatmap expression analysis using abiotic stress series from BAR (Toufighi et al., 2005). Rows represent genes and columns represent experimental conditions. The colour at a point represents the log2 of the ratio of the average of replicate treatments relative to the average of corresponding controls. The scale is shown in the figures. Orange boxes: ADC2 overexpressing plants. Blue boxes: SAMDC1 overexpressing plants; yellow boxes: SPMS overexpressing plants.
Transcriptome analyses presented in this work show that alteration of PA levels alters expression profiles of a significant number of genes. The existence of a PA modulon expression system in plants could be one of the possible explanations for some of the transcriptional changes observed in PA accumulating plants. A PA modulon composed by several key transcriptions factors stimulated by PAs at the translational level, has been proposed in Escherichia coli (Igarashi and Kashiwagi, 2006) and yeast (Uemura et al., 2009), although no clear evidences have been found in plants. The existence of regulation at translation level by PAs has been demonstrated for SAMDC in Arabidopsis (Hanfrey et al., 2003). Further efforts should be made toward the identification of transcription factors controlling the expression of genes by PAs, to get insight into the potential existence of a PA modulon in plants.
Some of the transcriptional changes observed could be direct or indirect consequence of crosstalking between PAs with other signaling pathways including ROS signaling and ABA biosynthesis (Alcázar et al., 2010b). Indeed, there are previous evidences of crosstalk between PAs and ABA. Upregulation of PA biosynthesis genes ADC2, SPDS1, and SPMS and accumulation of Put under drought stress in Arabidopsis are ABA-dependent responses (Alcazar et al., 2006a). Evidence of crosstalk between Spm and ABA is also shown in SAMDC1 overexpressing Arabidopsis plants. These lines show elevated levels of ABA due to the induction of NCED3, a key enzyme involved in ABA biosynthesis (Marco et al., in press).
On the other hand, a possible link between PAs, Ca2+ homeostasis and stress responses has also been suggested (Alcázar et al., 2010a). Spm control of Ca2+ allocation through regulating Ca2+ permeable channels, including CAXs, has been described as a possible mechanism of action for the protective role of Spm against high salt and drought stress (Yamaguchi et al., 2006, 2007). Moreover, changes of free Ca2+ in the cytoplasm of guard cells are involved in stomatal movement that may explain drought tolerance induced by Spm. Furthermore, Ca2+ signaling genes are one of the gene categories mainly upregulated in Put and Spm-overproducer plants (Fig. 4). In addition, a Spm signaling pathway has been proposed to explain the role of enhanced PA accumulation observed during pathogen response in Arabidopsis (Mitsuya et al., 2009; Takahashi et al., 2003). This signaling pathway could function via the merged signal of Spm-activated Ca2+-influx and H2O2 produced by Spm-degradation by PA oxidases. Both processes are able to trigger mitochondrial disfunction, and cell death (Takahashi et al., 2003). Mitsuya et al. (2009) identified a set of genes commonly induced by Spm and Cucumber Mosaic virus (CMV) infection. Seven Spm-upregulated genes identified in this study are also overexpressed in the Spm-overproducer plants, including transcription factor AtbZIP60, and mitogen-activated protein kinase AtMAPK3. AtbZIP60 may control the expression of genes participating in protein folding and secretion, which may be required during CMV-elicited hypersentitive response (HR) (Iwata and Koizumi, 2005). AtMAPK3 is an ortholog of a wound-induced protein kinase (WIPK) whose expression is induced by Spm in tobacco (Takahashi et al., 2003).
Therefore, evidences shown above indicate that PAs could act as key regulatory molecules in both abiotic and biotic stress processes, acting by crosstalk with other stress related hormones like ABA, and mediating through Ca2+ signaling. Remarkably, Put accumulation in ADC2 overexpressor plants also dampened gibberellic acid (GA) biosynthesis, thus evidencing the occurrence of intricate crosstalks between Put, GAs and ABA (Alcazar et al., 2005; Cuevas et al., 2008). The nature of these crossregulations could be different depending on the PA involved.
In summary, accumulation of Put and/or Spm is capable of triggering activation of biotic/abiotic defense mechanisms. This evidence, in addition to the PA roles as antioxidant and structural stabilizers, provides promising applications of PA-content manipulation to crop breeding for enhanced stress tolerance (Alcázar et al. 2010a; Gill and Tuteja, 2010).
Supplementary Material
Acknowledgments
A.F. Tiburcio was supported by grant BIO2008-05493-CO2-01 (MICINN); F. Marco and P. Carrasco were funded by grant BIO2005-09252-C02-02 (MICINN). R. Alcázar, A.F. Tiburcio, and P. Carrasco also acknowledge grants-in-aid from ACTION COST FA0605.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Alcázar R. Garcia-Martinez J.L. Cuevas J.C. Tiburcio A.F. Altabella T. Overexpression of ADC2 in Arabidopsis induces dwarfism and late-flowering through GA deficiency. Plant J. 2005;43:425–436. doi: 10.1111/j.1365-313X.2005.02465.x. [DOI] [PubMed] [Google Scholar]
- Alcázar R. Cuevas J.C. Patron M. Altabella T. Tiburcio A.F. Abscisic acid modulates polyamine metabolism under water stress in Arabidopsis thaliana. Physiol Plant. 2006a;128:448–455. [Google Scholar]
- Alcázar R. Marco F. Cuevas J.C. Patron M. Ferrando A. Carrasco P., et al. Involvement of polyamines in plant response to abiotic stress. Biotechnol Lett. 2006b;28:1867–1876. doi: 10.1007/s10529-006-9179-3. [DOI] [PubMed] [Google Scholar]
- Alcázar R. Altabella T. Marco F. Bortolotti C. Reymond M. Koncz C., et al. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010a;231:1237–1249. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- Alcázar R. Planas J. Saxena T. Zarza X. Bortolotti C. Cuevas J., et al. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol Biochem. 2010b;48:547–552. doi: 10.1016/j.plaphy.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Bannenberg G. Martínez M. Hamberg M. Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44:85–95. doi: 10.1007/s11745-008-3245-7. [DOI] [PubMed] [Google Scholar]
- Bouchereau A. Aziz A. Larher F. Martin-Tanguy J. Polyamines and environmental challenges: recent development. Plant Sci. 1999;140:103–125. [Google Scholar]
- Carmona-Saez P. Chagoyen M. Tirado F. Carazo J.M. Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007:8. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J.C. Lopez-Cobollo R. Alcázar R. Zarza X. Koncz C. Altabella T., et al. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 2008;148:1094–1105. doi: 10.1104/pp.108.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J.C. Lopez-Cobollo R. Alcázar R. Zarza X. Koncz C. Altabella T., et al. Putrescine as a signal to modulate the indispensable ABA increase under cold stress. Plant Signal Behav. 2009;4:219–220. doi: 10.4161/psb.4.3.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. Tuteja N. Polyamines and abiotic stress tolerance in plants. Plant Signal Behav. 2010;5:26–33. doi: 10.4161/psb.5.1.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M.E. Marco F. Gómez E. Carrasco P. Blázquez M.J. Carbonell J., et al. Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis thaliana defense against Pseudomonas viridiflava. Plant Physiol. 2011;156:2266–2277. doi: 10.1104/pp.110.171413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppa M.D. Benavides M.P. Polyamines and abiotic stress: recent advances. Amino Acids. 2008;34:35–45. doi: 10.1007/s00726-007-0501-8. [DOI] [PubMed] [Google Scholar]
- Hanfrey C. Franceschetti M. Mayer M.J. Illingworth C. Elliott K. Collier M., et al. Translational regulation of the plant S-adenosylmethionine decarboxylase. Biochem Soc Trans. 2003;31:424–427. doi: 10.1042/bst0310424. [DOI] [PubMed] [Google Scholar]
- Hanzawa Y. Imai A. Michael A.J. Komeda Y. Takahashi T. Characterization of the spermidine synthase-related gene family in Arabidopsis thaliana. FEBS Lett. 2002;527:176–180. doi: 10.1016/s0014-5793(02)03217-9. [DOI] [PubMed] [Google Scholar]
- Igarashi K. Kashiwagi K. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem. 2006;139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- Iwata Y. Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowitz T. Kneifel H. Piotrowski M. Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett. 2003;544:258–261. doi: 10.1016/s0014-5793(03)00515-5. [DOI] [PubMed] [Google Scholar]
- Kakehi J.I. Kuwashiro Y. Niitsu M. Takahashi T. Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1342–1349. doi: 10.1093/pcp/pcn109. [DOI] [PubMed] [Google Scholar]
- Knott J.M. Romer P. Sumper M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007;581:3081–3086. doi: 10.1016/j.febslet.2007.05.074. [DOI] [PubMed] [Google Scholar]
- Marco F. Buso E. Gruissem W. Lafuente T. Carrasco P. A possible role for spermine signalling during abiotic stress. Plant Physiol. 2011 (in press). [Google Scholar]
- Mitsuya Y. Takahashi Y. Berberich T. Miyazaki A. Matsumura H. Takahashi H., et al. Spermine signaling plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus. J Plant Physiol. 2009;166:626–643. doi: 10.1016/j.jplph.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R. Carmona-Saez P. Vazquez M. Vicente C. Yang X. Tirado F., et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicot M. Minguet E.G. Ferrando A. Alcazar R. Blazquez M.A. Carbonell J., et al. A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell. 2002;14:2539–2551. doi: 10.1105/tpc.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M. Janowitz T. Kneifel H. Plant C–N hydrolases and the identification of a plant N-carbamoylputrescine amidohydrolase involved in polyamine biosynthesis. J Biol Chem. 2003;278:1708–1712. doi: 10.1074/jbc.M205699200. [DOI] [PubMed] [Google Scholar]
- Schwartz S.H. Tan B.C. Gage D.A. Zeevaart J.A.D. McCarty D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Takahashi Y. Berberich T. Miyazaki A. Seo S. Ohashi Y. Kusano T. Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. Plant J. 2003;36:820–829. doi: 10.1046/j.1365-313x.2003.01923.x. [DOI] [PubMed] [Google Scholar]
- Thimm O. Blasing O. Gibon Y. Nagel A. Meyer S. Kruger P., et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tiburcio A.F. Altabella T. Ferrando A. Plant with resistance to low temperature and method of production thereof. Spanish patent application WO2010/004070. 2009.
- Toufighi K. Brady S.M. Austin R. L.E. Provart N.J. The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Tusher V.G. Tibshirani R. Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T. Higashi K. Takigawa M. Toida T. Kashiwagi K. Igarashi K. Polyamine modulon in yeast—stimulation of COX4 synthesis by spermidine at the level of translation. Int J Biochem Cell Biol. 2009;41:2538–2545. doi: 10.1016/j.biocel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Urano K. Yoshiba Y. Nanjo T. Igarashi Y. Seki M. Sekiguchi F., et al. Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ. 2003;26:1917–1926. [Google Scholar]
- Usadel B. Nagel A. Thimm O. Redestig H. Blaesing O. Palacios-Rojas N., et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.B. Malmberg R.L. Regulation of Arabidopsis thaliana (L.) heynh arginine decarboxylase by potassium deficiency stress. Plant Physiol. 1996;111:1077–1083. doi: 10.1104/pp.111.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.W. Yu W. Galloway G.L. Malmberg R.L. Isolation and characterization of a second arginine decarboxylase cDNA from Arabidopsis (Accession No. AF009647) Plant Physiol. 1997;114:1569. [Google Scholar]
- Yamaguchi K. Takahashi Y. Berberich T. Imai A. Miyazaki A. Takahashi T., et al. The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 2006;580:6783–6788. doi: 10.1016/j.febslet.2006.10.078. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K. Takahashi Y. Berberich T. Imai A. Takahashi T. Michael A.J., et al. A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem Biophys Res Commun. 2007;352:486–490. doi: 10.1016/j.bbrc.2006.11.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.