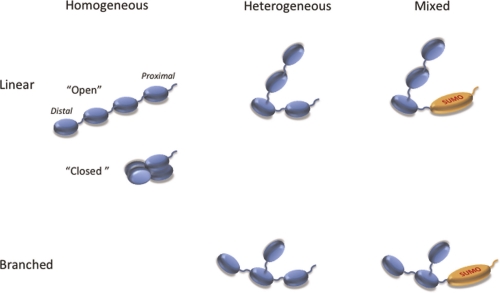
Figure 1. Architecture of polyubiquitin chains.
Unanchored chains are shown here. These are often found attached to a target protein via an isopeptide bond between the free C-terminus of the proximal ubiquitin and an amino group on the target protein. Chains can be “linear” (no more than one amino group of each ubiquitin is linked to another ubiquitin) or “branched” (at least one ubiquitin is attached to other ubiquitins via two or more different amino groups). The ubiquitin containing the free C-terminus is the “proximal” subunit and the ubiquitin(s) lacking any amino group linkage is a “distal subunit”. The polymers are referred to as “homogeneous” if all linkages use the same amino group on each ubiquitin. They are termed “heterogeneous” if ubiquitin linkages involve different amino groups on different subunits. Chains containing other ubiquitin-like proteins are called “heterologous” or “mixed”. The type of linkage is designated by the identity of the amino group used, i.e. Met1-, Lys6-, Lys11-, Lys27-, Lys29-, Lys33-, Lys48-, Lys63-linked. Note that Met1-linked ubiquitin is identical to the proprotein product of the polyubiquitin gene and has often been referred to as linear polyubiquitin. We propose the Met1-linked nomenclature to avoid the confusion with unbranched chains described above. In the “open” conformations the hydrophobic patch of ubiquitin is exposed, while in the “closed” conformations this patch is buried by ubiquitin-ubiquitin interactions. Abbreviations: SUMO, small ubiquitin-like modifier.