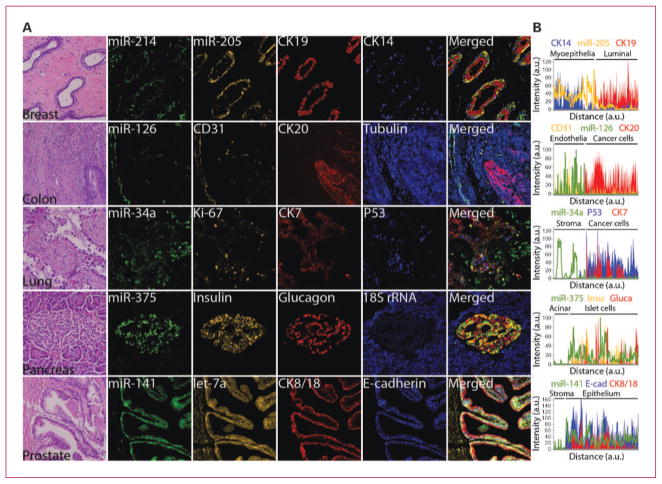
Fig. 2.
Codetection of miRNAs with cell type–specific and prognostic protein markers. A, serial FFPE tissue sections of the indicated organs were stained with H&E to reveal histologic features or subjected to ISH assay using FAM2×-tagged probes against miR-34a, miR-126, miR-141, miR-214, and miR-375 mixed with Bio2×-tagged probe against 18S rRNA, BrdU2×-tagged probe against miR-205, or DIG2×-tagged probe against let-7a. miRNA signals were revealed by sequential TSA reactions with FITC-tyramine (green for FAM2× probes), rhodamine-tyramine (orange for BrdU2× and DIG2× probes), and AMCA-tyramine (blue for Bio2× probe) substrates. After HIER as needed (Supplementary Table S2), expression of the indicated proteins was revealed by sequential TSA reactions with Dylight594-tyramine or Dylight680-tyramine (red) and AMCA-tyramine (blue) substrates, each TSA reaction was preceded by incubation with specific antibodies against each protein and appropriate anti-host species/HRP antibody; except for insulin expression, which was revealed by an anti–guinea pig secondary antibody conjugated to Cy3 (orange), obviating the need for the TSA step. B, line profile analysis was used to quantitate the intensity of RNA or protein expression. Background intensity was subtracted and intensity values were normalized setting the point with maximum intensity to 100 and calculating other values in relation to this reference. miRNA expression pattern was plotted as a line; independently, expression patterns of each protein were plotted as stacked areas. Displayed images were modified by optimizing the contrast with the process enhancement function from cellSens software package (Olympus), please see Supplementary Fig. S3 for raw fluorescent images and details on signal analysis.