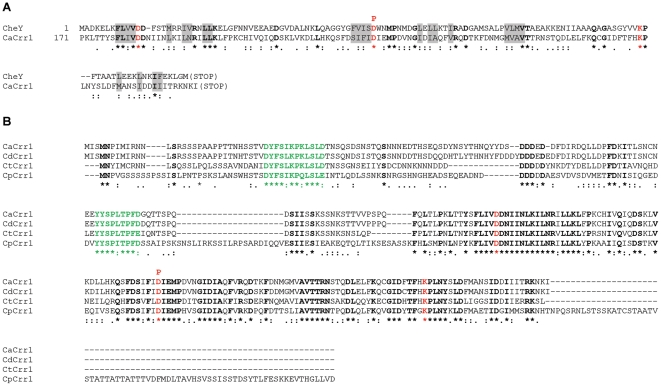
Figure 1. Sequence analysis of the response regulator protein Crr1 in C. albicans and identification of homologues in other diploid members of the Candida CTG clade.
(A) Clustal alignment of the receiver domain located in the C-terminal region of Crr1 (orf19.5843) of C. albicans (CaCrr1) and the CheY response regulator protein of Escherichia coli which essentially consists of a receiver domain. Residues that are identical between the receiver domains are indicated by bold, the aspartate and lysine residues conserved in all receiver domains are shown in red bold, and the aspartate residue which is predicted to be phosphorylated by two component signal transduction by a bold red “P”. Hydrophobic residues that are components of the hydrophobic core of CheY are indicated by grey shading. Note that the homology between the receiver domains extends to the replacement of amino acids with others with similar chemical properties. A colon indicates a highly similar substitution and a full stop a similar substitution. (B) Clustal alignment of potential Crr1 homologues in C. albicans (CaCrr1), C. dubliniensis (CD36_30940; CdCrr1), C. tropicalis (CTRG_00590; CtCrr1), and C. parapsilosis (CPAG_04104; CpCrr1). Residues shared by all four proteins are highlighted as described in (A) above. Two conserved regions were identified (green bold) that lie N-terminal to the potential receiver domain in each protein. The predicted protein sequences of the Crr1 homologues in the diploid members of the Candida CTG clade were obtained by BLAST analyses at the C. albicans genome web site (http://candidagenome.org/).