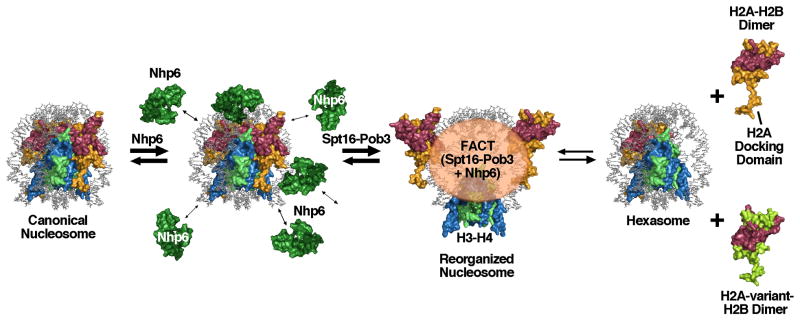
Figure 2.
Model for nucleosome reorganizing activities
Nhp6 binds to DNA at the surface of a canonical nucleosome, bending the DNA and destabilizing histone-DNA contacts [39]. Repeated binding-bending-release cycles within a short window of time render the nucleosome less stable and more suitable for reorganization by Spt16-Pob3. Reorganization involves disruption of the contacts between H2A–H2B and H3–H4, but the components of the nucleosome remain tethered together by FACT in a form in which the DNA is largely accessible. This dynamic form loses H2A–H2B dimers at a higher frequency than canonical nucleosomes do, but in the “global accessibility” model, dimer loss is an optional outcome of reorganization, not an obligate product. In the “dimer displacement” model FACT activity proceeds directly to displacement of an H2A–H2B dimer to form a hexasome. FACT might promote insertion of variant forms of H2A–H2B by chaperoning them and swapping them into either the reorganized form or hexasomes. The extended C-terminal tail of H2A that forms the docking domain that is partially responsible for dimer-tetramer stability in the core octamer is labeled; this domain may retain contact with H3–H4 during reorganization by bending at the point where it reaches the globular domain, unlike the rigid-body movement modeled here.