ABSTRACT
In Gram-negative bacteria, the Lol and Bam machineries direct the targeting of lipidated and nonlipidated proteins, respectively, to the outer membrane (OM). Using Pseudomonas aeruginosa strains with depleted levels of specific Bam and Lol proteins, we demonstrated a variable dependence of different OM proteins on these targeting pathways. Reduction in the level of BamA significantly affected the ability of the β-barrel membrane protein OprF to localize to the OM, while the targeting of three secretins that are functionally related OM proteins was less affected (PilQ and PscC) or not at all affected (XcpQ). Depletion of LolB affected all lipoproteins examined and had a variable effect on the nonlipidated proteins. While the levels of OprF, PilQ, and PscC were significantly reduced by LolB depletion, XcpQ was unaffected and was correctly localized to the OM. These results suggest that certain β-barrel proteins such as OprF primarily utilize the complete Bam machinery. The Lol machinery participates in the OM targeting of secretins to variable degrees, likely through its involvement in the assembly of lipidated Bam components. XcpQ, but not PilQ or PscC, was shown to assemble spontaneously into liposomes as multimers. This work raises the possibility that there is a gradient of utilization of Bam and Lol insertion and targeting machineries. Structural features of individual proteins, including their β-barrel content, may determine the propensity of these proteins for folding (or misfolding) during periplasmic transit and OM insertion, thereby influencing the extent of utilization of the Bam targeting machinery, respectively.
IMPORTANCE
Targeting of lipidated and nonlipidated proteins to the outer membrane (OM) compartment in Gram-negative bacteria involves the transfer across the periplasm utilizing the Lol and Bam machineries, respectively. We show that depletion of Bam and Lol components in Pseudomonas aeruginosa does not lead to a general OM protein translocation defect, but the severity (and therefore, Lol and Bam dependence), varies with individual proteins. XcpQ, the secretin component of the type II secretion apparatus, is translocated into the OM without the assistance of Bam or Lol machineries. The hypothesis that XcpQ, after secretion across the cytoplasmic membrane, does not utilize the OM targeting machineries was supported by demonstrating that in vitro-synthesized XcpQ (but not the other P. aeruginosa secretins) can spontaneously incorporate into lipid vesicles. Therefore, the requirement for ancillary factors appears to be, in certain instances, dictated by the intrinsic properties of individual OM proteins, conceivably reflecting their propensities to misfold during periplasmic transit.
Introduction
The outer membrane (OM) of Gram-negative bacteria is the outermost membrane barrier and is separated from the cytosol by the cytoplasmic or inner membrane (IM), the peptidoglycan layer, and the periplasm. Like the IM, the OM contains phospholipids but has a unique lipid component, the lipopolysaccharide (LPS), located exclusively in the outer leaflet of the bilayer. It also contains outer membrane proteins (OMPs), some of which are lipidated. In addition to contributing to the maintenance of cellular integrity, the OM is a key mediator of bacterial interaction with the environment and serves as an anchor for multicomponent systems involved in motility and protein secretion and in the import and export of small molecules. Recent work by several laboratories has shown that assembly of OMPs lacking or containing an acyl modification follows dedicated targeting pathways. Newly synthesized nonlipidated, β-barrel OMPs are translocated across the IM by the Sec machinery. Following the removal of the N-terminal signal sequence by the signal peptidase, the proteins are released into the periplasm and are shuttled in an unfolded state to the OM by chaperones SurA, Skp, and DegP. The assembly of β-barrel OM proteins is catalyzed by a machinery called the Bam complex that consists of a β-barrel membrane protein, BamA, and four lipoproteins, BamB, BamC, BamD, and BamE (1–3). BamA was first discovered (and designated Omp85) in Neisseria meningitidis, where it participates in the terminal steps in the OM insertion of the PorA and PorB porins, the FrpB siderophore receptor, and PilQ, the secretin component of the type IV pilus biogenesis apparatus (4). In N. meningitidis, the assembly of OMPs requires an additional component, RmpM, a periplasmic protein associated with the OM through binding to integral OMPs (5). The Bam complex is also important for correct localization of so-called autotransporters, proteins that utilize a C-terminal domain (“β domain”) to translocate an N-terminal extracellular domain (“passenger domain”) across the OM in a number of bacteria (6–9). The broad distribution of proteins whose assembly depends on the Bam complex suggested that this pathway represents a universal mechanism for translocating nonlipidated proteins with β-barrel structures across the periplasm into the OM. However, Collin et al. demonstrated that, in contrast to the porins OmpA and LamB, multimerization and OM association of the secretin PulD, a component of the type II secretion system (T2SS) in Klebsiella oxytoca, in which the presence of β strands remains uncertain because of the lack of structural information, occurred independently of BamA (10). Whether differences in the results obtained with the related PilQ and PulD proteins reflect a species-specific phenomenon and whether certain OMPs are assembled via another, as-yet-unidentified pathway remain to be determined.
The pathway for localization of outer membrane lipoproteins has been described in detail mostly for Escherichia coli (11, 12). The N-terminal signal peptides of lipoproteins enable their recognition by and transport through the Sec translocon as prolipoprotein precursors. Processing of the IM-bound precursor occurs through a series of sequential enzymatic events that involve Lgt, LspA, Lnt, and an N-terminal cis element called a lipobox. First, Lgt catalyzes addition of N-acyl-diacylglyceryl to the cysteine that is the last amino acid of the lipobox and the first amino acid of mature lipoprotein. This step is followed by cleavage of the signal peptide by the LspA signal peptidase. The Lnt protein completes the formation of the mature lipoprotein by catalyzing the aminoacylation of the diacylglycerylcysteine (for a review, see reference 12). A mature lipoprotein possessing an amino acid residue other than aspartate as the second residue (the “+2” sorting rule) is destined for the OM. These lipoproteins interact with the LolCDE complex in the IM, which then, upon ATP hydrolysis, transfers the lipoprotein to the periplasmic chaperone LolA, which directs them to the OM. At that point, LolB (itself a lipoprotein) serves as a receptor for the translocated lipoproteins, facilitating their insertion into the OM. Since four of the components of the E. coli Bam complex are lipoproteins, the secretion of nonlipidated proteins depends, indirectly, on the functional Lol pathway.
In order to define the range of substrates that share a common mechanism for localization, we selected three secretins of Pseudomonas aeruginosa, PilQ, XcpQ, and PscC, and determined the effects that reductions in the amounts of the BamA and LolB have on their translocation and assembly into the OM. PilQ, XcpQ, and PscC form multimeric ring-like structures in the OM and function during secretion of pilin subunits for assembly of type IV pili, secretion of substrates by the type II secretion system (T2SS), and translocation of protein substrates into host cells by the type III secretion system (T3SS), respectively (13). P. aeruginosa PilQ is closely related to N. meningitidis PilQ, while XcpQ is closely related to PulD. While the amounts of all three P. aeruginosa secretins were relatively unaffected or minimally affected by changes in BamA levels, the amounts of PilQ and PscC, but not XcpQ, were significantly reduced in the absence of LolB. We further show that in vitro-synthesized XcpQ, but not PilQ or PscC, assembles into liposomes. These findings raise the possibility that there exist multiple pathways and mechanisms for integration of specific proteins into the OM.
RESULTS
Depletion of BamA or LolB leads to an irreversible growth defect.
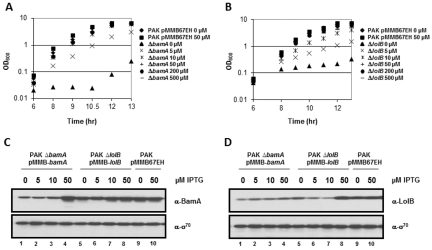
To assess the role of the Bam and Lol pathways in the targeting of various P. aeruginosa OMPs, we deleted the chromosomal bamA or lolB gene in a PAK strain that carries the corresponding genes in the pMMB67EH expression vector behind the tac promoter. The plasmid vectors in these strains, PAK ∆bamA (pMMB-bamA) (designated as the BamA-depletion strain) and PAK ∆lolB (pMMB-lolB) (designated as the LolB-depletion strain), also carry the gene for the LacI repressor, making the expression of the bamA and lolB genes dependent on the presence of the inducer isopropyl β-d-1-thiogalactopyranoside (IPTG) in the growth medium. When each strain was grown in Luria-Bertani (LB) media containing IPTG in a range of 0 to 500 µM, growth of the bacteria in the culture became dependent on the levels of the inducer, indicating that bamA and lolB are essential (Fig. 1A and B). Induction with 10 to 500 µM IPTG in the BamA-depletion strain yielded bacterial growth rates comparable to that of the PAK pMMB67EH wild-type (WT) strain, whereas the addition of 5 µM IPTG yielded a growth rate that lagged by approximately 3 to 4 doubling times. Without any induction (i.e., in the absence of IPTG), growth was severely attenuated in the BamA-depletion strain. A similar pattern of growth was observed for the LolB-depletion strain, which required at least 10 to 50 µM IPTG for growth close or comparable to that of WT PAK pMMB67EH. Differences in growth between the two strains carrying the bamA and lolB constructs were likely due to the requirement for certain levels of each product for the optimal functioning of the respective OM protein targeting apparatus. For subsequent experiments, unless indicated otherwise, cultures of PAK ∆bamA (pMMB-bamA) and PAK ∆lolB (pMMB-lolB) were supplemented with 5 µM and 50 µM IPTG for depleted and induced conditions, respectively.
FIG 1 .
The effect of BamA or LolB depletion on P. aeruginosa growth. Growth of P. aeruginosa PAK ∆bamA (pMMB-bamA) (A) and of P. aeruginosa PAK ∆lolB (pMMB-lolB) (B) in media containing various concentrations of IPTG. Growth of wild-type (WT) P. aeruginosa PAK containing the empty vector (pMMB67EH) either in the presence of 50 µM IPTG or without IPTG was also monitored. Immunoblots of cells grown in various concentrations of IPTG for 9 h and probed with antibodies against BamA (C) and LolB (D). Blots were also probed with antibodies against the σ70 subunit of RNA polymerase.
Immunoblotting was used to monitor the effect of IPTG on the levels of BamA and LolB. In each case, the decrease in growth correlated with a decrease in the level of BamA or LolB. At 50 µM IPTG, both BamA and LolB were overproduced approximately 2-fold compared to the WT control results (Fig. 1C and D), without having a deleterious effect on growth (Fig. 1A and B). While the depletion of BamA did not significantly affect the levels of LolB (lanes 1 to 4 in Fig. 1D), a noticeable decrease in the level of BamA was seen in a LolB depletion strain under conditions of IPTG limitation (lanes 5 to 8 in Fig. 1C). Interestingly, in both cases the growth defect was observed when the levels of BamA or LolB were reduced to approximately 25% of the WT level by limiting the IPTG concentration to 5 µM, and a more severe defect was seen in the absence of IPTG. The apparent IPTG-independent synthesis of BamA or LolB was presumably due to the leakiness of the tac promoter in pMMB67EH. These results suggest that the levels of BamA and LolB needed to sustain WT levels of growth cannot be reduced by more than 75% or else the ability of the bacteria to proliferate is impaired.
To assess whether depletion of either BamA or LolB is reversible, PAK ∆bamA (pMMB-bamA) and PAK ∆lolB (pMMB-lolB) were grown under conditions that resulted in a significant decrease in BamA and LolB levels (IPTG at 0 or 5 µM for 9 h) as well as in media containing 50 µM IPTG for full expression. Bacteria were then plated onto LB agar containing 50 µM IPTG, and viable bacteria were enumerated by determining the plating efficiency of the culture (i.e., comparing observed colony forming units [CFUs] to those expected based on normalized values for optical density at 600 nm [OD600]) (see Fig. S1 in the supplemental material). In each case, prior depletion of BamA and LolB before plating resulted in a significant loss of colony-forming ability, indicating that the observed impairment in the OMP transport (see below) led to irreversible damage and cell death.
Depletion of BamA or LolB affects the stability of the major β-barrel OMP OprF.
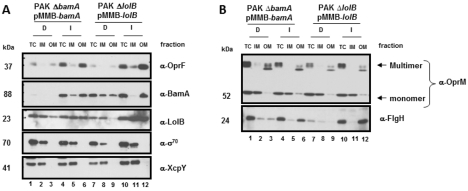
Whether the Bam and Lol pathways function in P. aeruginosa was examined by monitoring the OM localization of OprF, a well-characterized β-barrel OMP, as well as those of lipoproteins OprM and FlgH. Depleted (D) and inducing (I) conditions, controlled by the concentrations of IPTG at 0 and 50 µM IPTG, respectively, were used for the data presented in Fig. 2. Total cell (TC) levels of each protein in whole cells were determined, followed by fractionation of cell lysates into IM and OM compartments. The proper fractionation of the cells was monitored using antibodies against the cytoplasmic, σ70 subunit of RNA polymerase and the IM protein, XcpY. The depletion of BamA had a significant effect on the levels of detectable OprF in whole cells, suggesting that improper localization affects its stability, leading to degradation by periplasmic proteases (Fig. 2A, top panel, lanes 1 and 4). However, the residual OprF that was observed in these cells was correctly localized to the OM even under BamA-depletion conditions (Fig. 2A, top panel, lanes 3 and 6). BamA depletion did not affect oprF gene expression, since its transcript levels in the BamA-depleted and WT cells were similar (data not shown). A similar effect on the stability of OprF (and, to a lesser extent, on the stability of BamA) was seen in the LolB-depletion strain (Fig. 2A, the α-OprF and α-BamA blots, lanes 7 and 10). These results were not totally unexpected, since the Bam machinery contains four lipoproteins, and the effect of LolB depletion on the β-barrel OMP OprF might have been a consequence of inhibiting their trafficking to the OM via the Lol pathway.
FIG 2 .
Cellular localization of various OMPs in response to BamA or LolB depletion. Immunoblot analyses assessing the expression and localization of the OMPs OprF (A) and lipoproteins OprM and FlgH (B) were performed. The levels of individual proteins were determined in total cells (TC) and in the inner membrane (IM) and outer membrane (OM) fractions under BamA- or LolB-depleted (D) (0 µM IPTG) and induced (I; 50 µM IPTG) conditions. Antibodies specific to the σ70 and XcpY were used as loading and inner membrane markers, respectively. Equivalent amounts per milliliter with respect to the TC fraction were loaded for each lane. Samples were not heated to 100°C in SDS prior to SDS-PAGE analysis.
According to the results of examinations of total cell extracts or IM and OM fractions, BamA depletion did not significantly affect the total amounts of the lipoprotein OprM (monomers and slowly migrating multimers) (see lanes 1, 2, and 3 compared to lanes 4, 5, and 6 in Fig. 2B). If anything, in some experiments, there was a slight increase in the amount of this protein. However, the amount of OM-localized lipoprotein FlgH was reduced in BamA-depleted cells (compare lanes 3 and 6), suggesting that assembly of the flagellar apparatus containing this protein may depend on OM components translocated by the Bam apparatus. Contrary to expectations, the impairment of lipoprotein trafficking by depletion of LolB had only a modest effect on OprM: a lower level of OprM multimers was detected in the total cells whereas the amount of the same multimeric species in the OM was unaffected. The level of OprM monomers, located predominantly in the inner membrane, was also unaffected by LolB depletion. It is conceivable that the fraction of OprM not degraded in the periplasm in the absence of LolB is incorporated in the OM. In contrast, the total amount of FlgH was significantly lower when LolB was depleted (compare lanes 7 and 10 in Fig. 2B) and, since none was detected in the IM or OM, mistargeted FlgH was either degraded or lost from the periplasm during fractionation.
Effect of BamA and LolB depletion on the localization of P. aeruginosa secretins.
The availability of P. aeruginosa strains in which the levels of BamA or LolB can be manipulated allowed us to examine targeting of the P. aeruginosa PilQ, the ortholog of the BamA-dependent N. meningitidis pilin secretin. We also analyzed the OM localization of XcpQ and PscC, two secretins involved in distinct extracellular protein targeting machineries, the T2SS and T3SS, respectively. For these experiments, we modified PAK ∆bamA (pMMB-bamA) and PAK ∆lolB (pMMB-lolB) by introducing into the chromosome of each one a cassette containing the gene for the arabinose regulator (AraC) and the pBAD promoter oriented in such a way as to drive the expression of the pilQ, xcpQ, and pscC genes, as described in Materials and Methods. These strains allowed us to coordinate the depletion of BamA or LolB with arabinose-mediated induction of expression and assembly of the secretins into the OM.
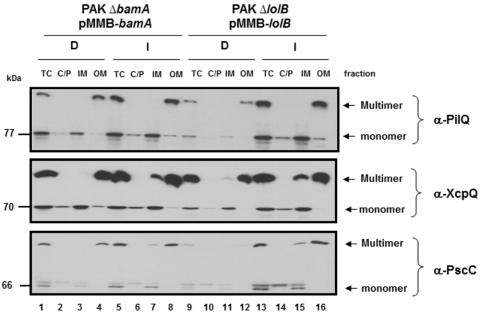
Comparison of the levels and IM and OM localization of PilQ, XcpQ, and PscC in BamA- and LolB-depletion strains is shown in Fig. 3. In cells with depleted BamA, the total levels of PilQ, XcpQ, and PscC monomers and multimers were virtually unaffected (XcpQ) or only marginally affected (PilQ and PscC) (compare lanes 1 and 5 in Fig. 3). The distribution of these proteins in the IM and OM was independent of BamA levels, and the total amount recovered in the membrane fractions corresponded to the amount of multimers detected in total cell extracts (Fig. 3, lanes 3 and 4 and 7 and 8). The pools of monomers in the soluble fractions (representing the cytoplasmic and periplasmic compartments) were also unaffected by the levels of BamA (Fig. 3, lanes 2 and 6). Therefore, unlike the OM localization of OprF, the insertion of the secretins PilQ and PscC does not appear to be strongly influenced by the levels of BamA. XcpQ appears to be completely independent of BamA.
FIG 3 .
Analysis of the localization of P. aeruginosa secretins in BamA- and LolB-depletion strains. The levels of PilQ, XcpQ, and PscC were determined by immunoblotting using antibodies raised against each secretin. P. aeruginosa PAK ∆bamA (pMMB-bamA) and PAK ∆lolB (pMMB-lolB), carrying an araCpBAD-pil, araCpBAD-xcpQ, or araCpBAD-exsA construct, were grown under depleted (D) conditions (5 µM IPTG) and induced (I) conditions (50 µM IPTG) for 8 h followed by induction of the secretin genes by addition of 0.02% (for pil and xcp) or 0.1% (for pscC) l-arabinose for 1 h. The amounts and localization of each secretin were analyzed in total cells (TC), and, following lysis, the combined cytoplasmic and periplasmic fractions (C/P) was separated from the cell envelopes by centrifugation. The cell envelopes were further fractioned into the inner membrane (IM) and outer membrane (OM) components. Equivalent amounts per milliliter with respect to the TC fraction were loaded for each lane. Samples were not heated to 100°C in SDS prior to SDS-PAGE analysis.
We next utilized the LolB-depletion strain to assess the contribution of this protein to the OM localization of the three P. aeruginosa secretins. In strains with depleted LolB, significantly reduced levels of PilQ and PscC monomers and multimers were observed (Fig. 3, top and bottom panels, lanes 9 and 13). In contrast, the effect of limiting LolB on the overall levels of XcpQ was minimal (Fig. 3, middle panel, lanes 9 and 13), and the OM localization and assembly of multimers was largely unaffected in the depleted strain (Fig. 3, middle panel, lanes 12 and 16). These results suggest that the OM lipoprotein assembly pathway makes a substantial contribution to the stability of PilQ and PscC and that failure to assemble into the OM very likely results in their degradation. Interestingly, LolB depletion had no effect on the levels, multimerization, and OM localization of XcpQ. These results demonstrate a significant role of correctly localized lipoproteins in the biogenesis of some, but not all, OM proteins.
Interaction of in vitro-synthesized secretins with liposomes.
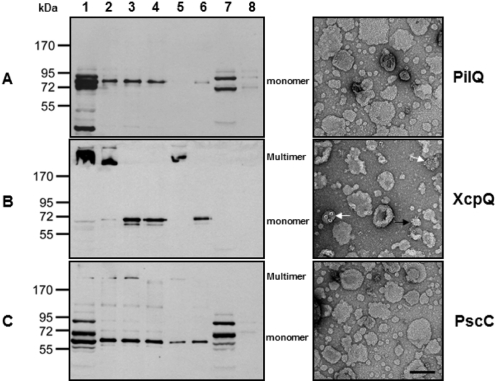
Previous studies have shown that PulD and OutD, secretins from the T2SSs of Klebsiella oxytoca and Erwinia chrysanthemi, respectively, can form stable multimers and insert spontaneously into liposomes when produced in vitro (14). To assess the requirement for exogenous factors during membrane assembly of the P. aeruginosa secretins, PilQ, XcpQ, and PscC (lacking their respective signal peptides) were synthesized using a coupled cell-free translation system in the presence of synthetic liposomes generated from a suspension of asolectin. Insertion of these proteins into the liposomes was monitored by centrifugation with and without treatment that is known to extract peripherally associated proteins (urea) or to dissociate secretin multimers completely (phenol). The results shown in Fig. 4 demonstrate that a significant fraction of in vitro-synthesized XcpQ, but not PilQ or PscC, formed high-molecular-mass multimers that associated with liposomes present during their synthesis (left panels, lanes 1 and 2). As observed for OutD, XcpQ multimers were dissociated into monomers by heat and phenol treatment (Fig. 4B, lanes 3 and 4). The XcpQ multimers remained associated with the liposomes after urea treatment, indicating that they were correctly inserted into the liposome bilayer (Fig. 4B, lane 5). Transmission electron microscopy (TEM) of XcpQ containing urea-treated liposomes clearly showed the presence of ring-like structures similar to those previously observed in vivo (Fig. 4B, right panel) (15). Analogous structures were not found to be associated with liposomes present after synthesis of PilQ and PscC (Fig. 4A and C, right panels). These results suggest that XcpQ differs from PilQ and PscC in its ability to form multimers spontaneously and insert into liposomes.
FIG 4 .
Analysis of in vitro-synthesized PilQ, XcpQ, and PscC insertion into liposomes. Secretins PilQ (A), XcpQ (B), and PscC (C), lacking signal sequences, were synthesized in vitro in the presence of asolectin liposomes. Samples were analyzed by SDS-PAGE and immunoblotting with the appropriate secretin-specific antisera and with or without prior urea treatment to dissociate peripherally associated proteins and phenol treatment to dissociate multimers. Equivalent amounts of each sample in relation to the amount of total reaction mixture represented in lane 1 were loaded. DNA encoding green fluorescent protein (Gfp) was used in a control reaction for proteins cross-reacting with the antisera. Lane 1, total in vitro reaction; 2, centrifuged liposomes; 3, liposomes heated to 100°C in SDS; 4, phenol-treated liposomes; 5, urea-washed liposomes; 6, urea-washed liposomes treated with phenol; 7, total in vitro reaction of Gfp control; 8, centrifuged liposomes of Gfp control. Transmission electron microscope images of negatively stained liposomes, after being washed with 4 M urea, are shown on the right in each panel. The white arrows indicate top views, and the black arrow indicates a side view. Bar, 100 nm.
DISCUSSION
According to the current state of knowledge, transport of proteins to and their insertion into the OM occurs by one of two main pathways, depending on their lipidation status. Nonlipidated β-barrel-containing proteins utilize a group of periplasmic chaperones and the five OM Bam proteins for their folding and insertion into the OM (2, 3). Peripheral OM lipoproteins are first lipidated in the IM and then shuttled to the OM through a pathway consisting of inner membrane components (LolCDE), a periplasmic protein (LolA), and an integral OM lipoprotein (LolB) (11, 12). The two processes are not entirely independent, since BamB, BamC, BamD, and BamE are all lipoproteins and, therefore, their targeting to the OM likely utilizes the Lol pathway. Furthermore, OM lipoproteins with transmembrane β-barrel structures presumably use elements from both Lol and Bam pathways for targeting and assembly.
In this study, we developed P. aeruginosa strains in which the levels of BamA and LolB could be controlled and then examined the consequences of their depletion for several OM proteins. An approximately 75% reduction of either BamA or LolB caused a dramatic decline in the cellular level of OprF, the β-barrel OM porin, presumably due to degradation of the misfolded periplasmic intermediates. These findings are consistent with similar experiments performed with P. aeruginosa and other microorganisms in which insufficient amounts of BamA destabilized and prevented trafficking of β-barrel proteins (7, 8, 16). The effect of BamA depletion on lipoproteins (Fig. 2B) was modest at most, but levels of the lipoprotein FlgH (and to a lesser extent, OprM) were significantly reduced as a consequence of LolB depletion. In each case, we observed that a fraction of the lipoproteins escaped proteolytic degradation in the periplasm and that those lipoproteins were correctly targeted into the OM. These findings further underscore the significant role of the OM targeting machineries in preventing misfolding and protection against degradation of their substrates.
The most intriguing finding from the analyses of the BamA- and LolB-depletion strains is that the levels and, therefore, by inference, the trafficking of P. aeruginosa secretins (PilQ, XcpQ, and PscC) were unaffected or slightly (albeit noticeably and reproducibly) affected when BamA levels were sufficiently low to compromise growth and severely reduce trafficking of OprF. While this result is in line with that reported for secretin PulD produced in E. coli, for which BamA-independent multimerization and the ability to form multimers when mistargeted to the inner membrane were previously reported (10, 17), it is in marked contrast to the reported BamA dependence of PilQ assembly in N. meningitidis (4). While these two PilQ secretins perform identical functions during the biogenesis of type IV pili in their respective organisms, their sequences are not highly conserved. In contrast to BamA depletion, LolB depletion affected the stability and localization not only of lipoproteins but also of the nonlipidated secretins PilQ and PscC (but not XcpQ). The effects of LolB depletion described above could be explained by a requirement for the lipoprotein (BamB to BamE) components of the Bam pathway. Alternatively, as-yet-uncharacterized Lol-dependent lipoproteins might play a role in stabilizing and/or targeting PilQ and PscC (and perhaps additional OM proteins). One such lipoprotein might be PilF (18, 19), the homologue of the N. meningitidis lipoprotein PilW, which is required for PilQ assembly in this bacterium (20).
It is also noteworthy that XcpQ can insert into liposomes present during in vitro synthesis, (this study), as can PulD and OutD (14), suggesting that assembly and insertion of these three secretins is independent of protein components in the outer membrane. In contrast, P. aeruginosa secretins PscC and PilQ (this study) and N. meningitidis PilQ (14) are unable to multimerize and insert into liposomes present during in vitro synthesis, again indicating that system-specific or species-specific soluble or membrane components are probably required in these cases.
One explanation for the differing requirements for Bam components for OMP trafficking and assembly could be that certain OMPs, such as secretins, do not require all or even any components of the Bam machinery for their trafficking and assembly. Moreover, based on our results, some lipoproteins, such as OprM, may have an only limited requirement for the components of the Lol pathway. It should also be noted that OprM is a structural and functional homologue of the nonlipidated E. coli protein TolC, the assembly and insertion of which is independent of the POTRA domain 1 of BamA that is required for the assembly of classical β-barrel proteins (21). A previous study assessing the requirement of the Bam pathway for extracellular localization of the E. coli hemoglobin protease, an autotransporter with a β-barrel translocator domain, showed that the BamB lipoprotein was dispensable for its insertion into and secretion across the OM (9). These observations further illustrate that the requirement for different components of the Bam apparatus for OM localization can differ from protein to protein, presumably according to their individual structures and organization. Thus, one explanation for the apparent selective utilization by different OMPs of the conserved OM lipoprotein localization pathway is that the lipidated Bam proteins function individually, allowing specific OMPs to engage them as they are needed, based on their intrinsic folding properties and ability to avoid degradation in the periplasm. For example, certain β-barrel proteins, such as OprF, would normally fully engage all of the periplasmic and OM components of the Bam and Lol pathways needed for its correct OM insertion and, in the absence of any one of these components (such as BamA), would be trapped in the periplasm in a misfolded form and degraded by the DegP protease. In contrast, other proteins targeted to the OM could follow a pathway that depends (partially or totally) on only a selection of the Bam components. We note that BamA, the Bam component studied here, is generally considered to be the core element of the Bam machinery, and it is not clear whether other Bam components would function in its absence.
The observations reported here provide support for the hypothesis that, depending on certain intrinsic properties of different OMPs, their utilization of broad-specificity OM localization factors may range from complete to nonexistent. Proteins with strong β-barrel structural features appear to be uniformly dependent on some form of the OM translocation system. In contrast, proteins that do not adopt this structural conformation may be targeted into the OM by an alternative machinery, recognizing a segment or a folded domain lacking a β-barrel fold. It is also conceivable that some proteins, such as XcpQ, are targeted into the OM without the assistance of any ancillary cellular components. The variable dependence of the different OMPs on general trafficking factors could be determined, in part, by the ability of these proteins to fold spontaneously into a protease-resistant conformation in the periplasm and by the presence of sequence or structural features that allow spontaneous insertion into the OM. Proteins that function as folded β-barrel structures could be more prone to misfolding during periplasmic translocation and OM insertion, thus increasing their dependence on the Bam machinery. Interestingly, the results of the present study of secretins indicate that requirements for OM localization and assembly factors differ even for related proteins within the same bacterial cell, in this case, P. aeruginosa. An example of the use of yet another pathway for secretin targeting in this bacterium was provided by studies of HxcQ, which is a lipidated secretin that probably uses the Lol pathway to reach the OM directly (22). Likewise, only a limited number of secretins, of which PulD is the best-studied example, use a lipoprotein (pilotin) to direct them to the outer membrane via the Lol pathway (23). Pilotins are not known to be involved in the assembly or function of XcpQ, but secretin chaperones PilP and PscW, whose genes are linked to pilQ and pscC, respectively, as well as PilF as mentioned above, might modulate their OM assembly. As discussed previously (24), the use of such a wide variety of targeting and assembly mechanisms for even closely related secretins almost certainly reflects the need to prevent the formation of mixed multimers that would likely be nonfunctional. This is particularly important in bacteria such as P. aeruginosa that possess multiple secretin-dependent protein secretion machineries. Therefore, the results presented here provide an impetus for further investigation into targeting of different OM proteins in a range of taxonomically unrelated microorganisms and with a range of related OMPs, which should shed light on the finer features and diversity of protein trafficking mechanisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed and described in Table S1 and Text S1 in the supplemental material. Typically, unless otherwise noted, bacterial cultures were grown in Luria-Bertani (LB) broth at 37°C. Where appropriate, antibiotics were added at the following concentrations: carbenicillin at 50 µg/ml, gentamicin at 15 µg/ml (E. coli) or 75 µg/ml (P. aeruginosa), kanamycin at 50 µg/ml, and 5-chloro-2-(2,4-dichlorophenoxy)phenol (Irgasan) at 25 µg/ml.
Growth analysis of depletion mutants.
Overnight cultures were washed three times and inoculated at about 105 CFU/ml into the appropriate medium used for BamA- or LolB-depleted (low IPTG) or induced (high IPTG) conditions. The growth of each culture was assessed by measuring the optical density at 600 nm (OD600) at the indicated time points. To determine the mechanism of cell death in the absence of the relevant OMP assembly proteins (i.e., BamA or LolB), an aliquot from each culture was serially diluted in LB broth and then plated on LB agar plates containing 50 µM IPTG to fully induce expression of BamA or LolB. After an overnight incubation at 37°C, the numbers of CFU per milliliter were enumerated. The plating efficiency was calculated as the percentage of the measured CFU versus estimated CFU per milliliter.
Cell fractionation and immunoblotting.
Bacteria were grown as described above to 9 h postinoculation, and the cells were harvested by centrifugation at 4°C. The pellets were resuspended in lysis buffer (20 mM Tris-Cl [pH 7.5], 0.1 M NaCl, 1 mM EDTA, 1× Roche Complete protease inhibitor cocktail, lysozyme [0.5 mg/ml]) to a concentration of 2 × 1010 CFU/ml. After lysis by sonication, the cellular debris was cleared by centrifugation (10,000 × g, 10 min, 4°C). One half of the cell lysate (250 µl) from each sample was saved as the total cell (TC) fraction, and the remaining half was further fractioned by ultracentrifugation (200,000 × g, 1 h, 4°C). The supernatant constituted the soluble fraction (cytoplasmic and periplasmic contents). The pellet was resuspended in 250 µl of inner membrane extraction buffer (20 mM Tris-Cl [pH 7.5], 0.2% sodium lauroyl sarcosinate [Sarkosyl; Fluka]) and incubated on ice for 30 min. After a second ultracentrifugation, the supernatant was saved as the inner membrane fraction, whereas the pellet (outer membrane fraction) was resuspended in 250 µl of buffer (20 mM Tris-Cl [pH 7.5], 0.1% sodium dodecyl sulfate [SDS]). Equal volumes of each cell fraction were separated on an SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a polyvinylidene difluoride (PVDF) membrane, which was blocked overnight at 4°C with 5% nonfat milk in TBST buffer (25 mM Tris-Cl [pH 7.5], 137 mM NaCl, 3 mM KCl, 0.1% Tween 20). The membranes were probed with specific primary polyclonal antibody diluted in TBST buffer for 2 h at 25°C and then washed with TBST buffer 6 times for 5 min each time. The membranes were further incubated (for 2 h at 25°C) with the secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit antibody; Bio-Rad) in 5% nonfat milk-TBST buffer and washed. In the instances where monoclonal antibodies were used, the solution consisted of horseradish peroxidase goat anti-mouse as the secondary antibody. Signal detection was achieved with Supersignal West Pico chemiluminescent substrates (Pierce). All of the OMP-specific polyclonal antibodies used for the experiments were from the S. Lory laboratory collection.
In vitro protein synthesis.
To test for in vitro multimerization, each secretin (PilQ, XcpQ, or PscC) was synthesized by using a rapid transcription-translation system 100 E. coli HY kit (5-Prime) in the presence of 100 µg of asolectin liposomes (Sigma) and 0.5 µg of vector DNA at 30°C, as previously described (14). To check for multimer assembly and insertion into liposomes, the reaction mixture was diluted in phosphate-buffered saline (PBS) before centrifugation using a Beckman TLA-55 rotor at 23,000 × g for 10 min at 20°C. The pellet was resuspended in the same buffer, and aliquots of it and of the supernatant were either heated or treated with phenol and then analyzed by immunoblotting. A further aliquot of the resuspended pellet was centrifuged and resuspended in 4 M urea in the same buffer, incubated for 15 min at room temperature, and centrifuged again. The urea-washed pellet was resuspended, and an aliquot of both fractions was prepared for analysis as described above.
Electron microscopy.
Liposomes were centrifuged at 23,000 × g for 10 min, resuspended in 4 M urea for 15 min to dissociate liposomes and loosely bound material, diluted to 1 M urea by addition of TN buffer, centrifuged at 250,000 × g for 30 min, and resuspended in 20 mM Tris containing 250 mM NaCl (TN buffer). For imaging of negatively stained samples, aliquots of 5 µl were adsorbed on carbon film-coated copper grids, washed with 10 droplets of pure water, and subsequently stained with 2% uranyl-acetate. Images were recorded using a 2 K × 2 K charge-coupled-device (CCD) camera (Veleta; Olympus Company) at 92,000 × magnification and a Philips CM10 TEM operating at 80 kV.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download Text S1, DOCX file, 0.1 MB.
Strains and plasmids.
Irreversibility of BamA and LolB depletion. P. aeruginosa PAK (pMMB67EH), PAK ∆bamA(pMMB-bamA), and PAK ∆lolB (pMMB-lolB) were grown using various IPTG concentrations (0, 5, or 50 µM) for 9 h and then plated on media with 50 µM IPTG (inducing conditions). CFUs were enumerated after an overnight incubation, and the plating efficiency was calculated as the percentage of the actual versus estimated CFU per milliliter. Download Figure S1, EPS file, 0.5 MB.
ACKNOWLEDGMENTS
H. H. Hoang was supported by a Cystic Fibrosis Foundation postdoctoral fellowship, and N. N. Nickerson was supported by a postdoctoral fellowship from the Canadian Louis Pasteur Foundation. This study was also supported by a research grant from the Cystic Fibrosis Foundation to S. Lory and by ANR grant ANR-09-BLAN-0291 to A. P. Pugsley.
Footnotes
Citation Hoang HH, et al. 2011. Outer membrane targeting of Pseudomonas aeruginosa proteins shows a variable dependence on the components of Bam and Lol machineries. mBio 2(6):00246-11. doi:10.1128/mBio.00246-11.
REFERENCES
- 1. Reumann S, Davila-Aponte J, Keegstra K. 1999. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc. Natl. Acad. Sci. U. S. A. 96:784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu T, et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]
- 3. Sklar JG, et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265 [DOI] [PubMed] [Google Scholar]
- 5. Volokhina EB, Beckers F, Tommassen J, Bos MP. 2009. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191:7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ieva R, Bernstein HD. 2009. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 106:19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bodelón G, Marín E, Fernández LA. 2009. Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of intimin from enteropathogenic Escherichia coli strains. J. Bacteriol. 191:5169–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jain S, Goldberg MB. 2007. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189:5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sauri A, et al. 2009. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155:3982–3991 [DOI] [PubMed] [Google Scholar]
- 10. Collin S, Guilvout I, Chami M, Pugsley AP. 2007. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol. Microbiol. 64:1350–1357 [DOI] [PubMed] [Google Scholar]
- 11. Tokuda H. 2009. Biogenesis of outer membranes in Gram-negative bacteria. Biosci. Biotechnol. Biochem. 73:465–473 [DOI] [PubMed] [Google Scholar]
- 12. Tokuda H, Matsuyama S. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1694 (1-3):IN1–IN9 [PubMed] [Google Scholar]
- 13. Bitter W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179:307–314 [DOI] [PubMed] [Google Scholar]
- 14. Guilvout I, et al. 2008. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J. Mol. Biol. 382:13–23 [DOI] [PubMed] [Google Scholar]
- 15. Bitter W, Koster M, Latijnhouwers M, De Cock H, Tommassen J. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209–219 [DOI] [PubMed] [Google Scholar]
- 16. Tashiro Y, et al. 2008. Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol. 190:3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. 2006. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 25:5241–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim K, et al. 2006. Crystal structure of PilF: functional implication in the type 4 pilus biogenesis in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 340:1028–1038 [DOI] [PubMed] [Google Scholar]
- 19. Koo J, et al. 2008. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190:6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carbonnelle E, Hélaine S, Prouvensier L, Nassif X, Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64 [DOI] [PubMed] [Google Scholar]
- 21. Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77:1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viarre V, et al. 2009. HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J. Biol. Chem. 284:33815–33823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collin S, Guilvout I, Nickerson NN, Pugsley AP. 2011. Sorting of an integral outer membrane protein via the lipoprotein-specific Lol pathway and a dedicated lipoprotein pilotin. Mol. Microbiol. 80:655–665 [DOI] [PubMed] [Google Scholar]
- 24. Guilvout I, Nickerson NN, Chami M, Pugsley AP. 2011. Multimerization-defective variants of dodecameric secretin PulD. Res. Microbiol. 162:180–190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download Text S1, DOCX file, 0.1 MB.
Strains and plasmids.
Irreversibility of BamA and LolB depletion. P. aeruginosa PAK (pMMB67EH), PAK ∆bamA(pMMB-bamA), and PAK ∆lolB (pMMB-lolB) were grown using various IPTG concentrations (0, 5, or 50 µM) for 9 h and then plated on media with 50 µM IPTG (inducing conditions). CFUs were enumerated after an overnight incubation, and the plating efficiency was calculated as the percentage of the actual versus estimated CFU per milliliter. Download Figure S1, EPS file, 0.5 MB.