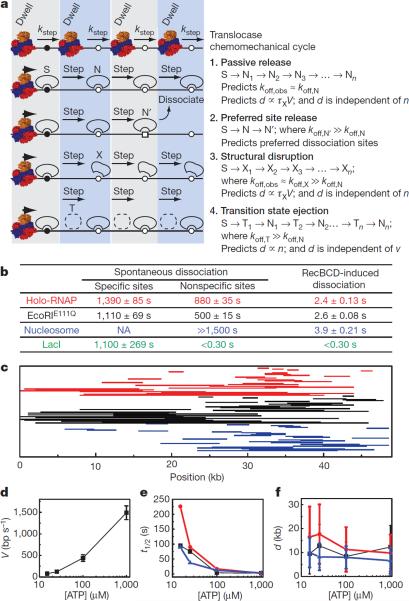
Figure 4.
Protein displacement mechanisms. a, Models for protein displacement. b, Spontaneous dissociation versus RecBCD-induced dissociation as determined from single exponential fits to dissociation data ± s.d. Values for LacI nonspecific and RecBCD-induced dissociation represent upper bounds. Data are color-coded for each different protein, as indicated. c, Representative sliding trajectories. Each line corresponds to the collision (right endpoint) and dissociation (left endpoint) for single proteins. d, RecBCD velocity (mean ± s.d.) at varying [ATP]. e, Protein t1/2 at varying [ATP]. Error was ≤5.6% of the reported values. f, Pushing distances (mean ± s.e.m.) at different ATP concentrations.