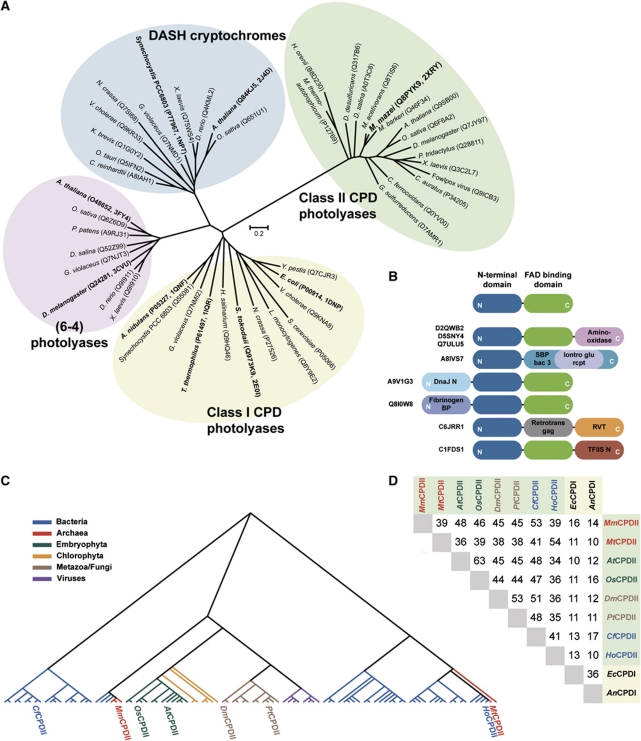
Figure 1.
Phylogenetic analysis of the class II photolyase subfamily. (A) The class II CPD photolyase subfamily forms a distinct cluster of light-dependent DNA-repair enzymes. The scale bar of the unrooted phylogenetic tree indicates amino-acid substitutions per site. (B) Several class II photolyases (UniProt identifiers depicted at left) show various kinds of domain fusions at the N- or C-terminal subdomains (blue and green). (C) Dendrogram of 98 non-redundant members of the class II photolyase subfamily. (D) Sequence identities of prominent members of class II and class I CPD photolyases. Class II CPD photolyases: Methanosarcina mazei (MmCPDII), Methanobacterium thermoautotrophicum (MtCPDII), Arabidopsis thaliana (AtCPDII), Oryza sativa (OsCPDII), Drosophila melanogaster (DmCPDII), Potorous tridactylus (PtCPDII), Chlorobium ferrooxidans (CfCPDII) and Halothermothrix orenii (HoCPDII); Class I CPD photolyases: Escherichia coli (EcCPDI) and Anacystis nidulans (AnCPDI). Phylogenetic analysis of class II photolyases is summarized in Supplementary data.