Figure 3.
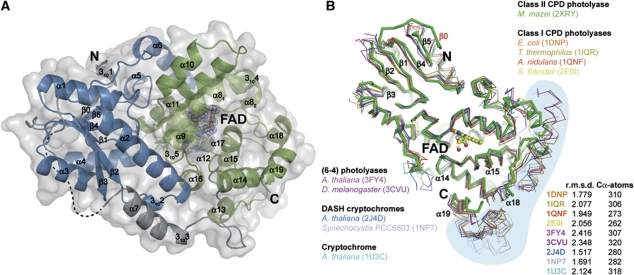
Overall structure of the M. mazei class II photolyase and structural comparison of members of the photolyase-cryptochrome family. (A) The N-terminal DNA-photolyase domain is shown in blue. C-terminal FAD-binding domain (green) contains the catalytic cofactor FAD (yellow) with SigmaA-weighted 2Fobs−Fcalc electron density contoured at 1σ. Domains are connected by a linker (grey) containing an α-helix (α7) with a 310-helical extension (3103). Not defined part of the linker by electron density is illustrated with a dashed line. Colouring of domain limits corresponds to Pfam database (Finn et al, 2010). (B) The M. mazei class II photolyase reveals a common overall fold to superimposed structures of the photolyase-cryptochrome family.