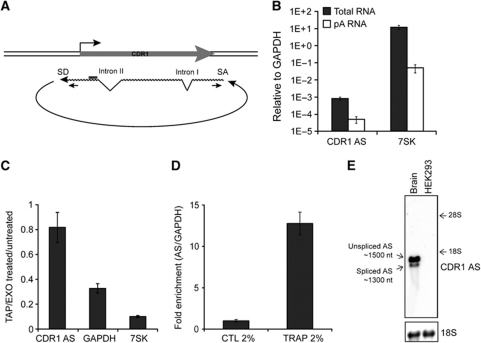
Figure 2.
Characterization of the circular CDR1 antisense RNA. (A) Schematic illustration of the circular antisense RNA including the NAS splice donor (SD) and splice acceptor (SA) sites and the position of the NAS-specific primer set. (B) qRT–PCR for total or poly(A)-enriched RNA from HEK293 using primers specific for the NAS RNA. 7SK is included as a poly(A) tail-deficient control RNA. (C) NAS-specific qRT–PCR for RNA treated with TAP (Tobacco Acid Pyrophosphatase) and EXO (Terminator 5′ Phosphate-Dependent Exonuclease) normalized to untreated RNA. GAPDH mRNA and 7SK RNA are included as controls for TAP and EXO efficiencies. (D) Total HEK293 RNA was mixed with solidifying agarose and subjected to electrophoresis. In this system, circular RNA species will be physically trapped, whereas linear RNAs remain free to migrate. A no-electrophoresis control was included to determine gel-extraction efficiency, and −fold enrichment in CDR1 AS was quantified by NAS-specific qRT–PCR relative to total INPUT RNA and normalized to GAPDH. (E) Agarose northern blot with 2 μg RNA from HEK293 cells or human brain showing distinct AS migration using a CDR1 NAS-specific probe (top panel). Unspliced and spliced AS represent splicing of canonical intron II (see A). The approximate RNA sizes were deduced based on ribosomal RNA migration. 18S (bottom panel) serves as loading control.