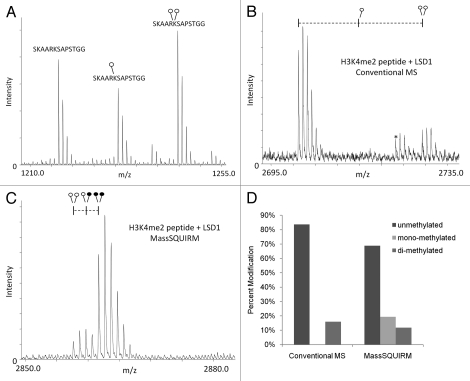
Figure 3.
MassSQUIRM accurately corrects for differences in ionization. (A) Equimolar amounts of an H3K27 peptide with 0, 1 or 2 methyl groups were mixed and analyzed by MALDI mass spectrometry. The un-, mono- and di-methylated peptides ionize at different efficiencies. (B) A synthetic peptide representing H3K4me2 was incubated with the histone demethylase LSD1 and the resulting peptides were analyzed by conventional mass spectro-metry. (C) The same reaction was performed as in (B) but peptides were analyzed using MassSQUIRM. (D) MassSQUIRM allows quantification of mono-methyl peptide species that was omitted in conventional mass spectrometry analysis due to differential ionization seen in (A). Open circles indicate light methylation while closed circles indicate heavy methylation.