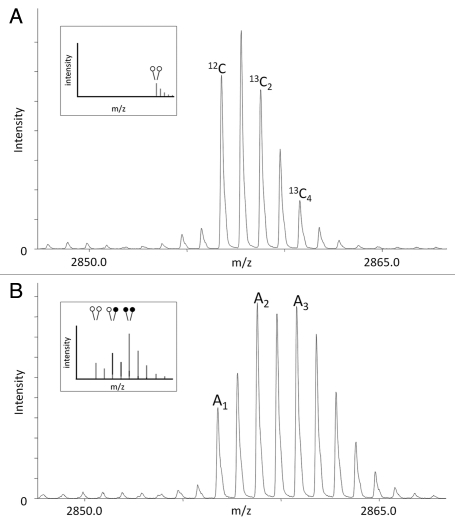
Figure 4.
MassSQUIRM can be used to successfully quantify differentially modified peptides. (A) A di-methylated synthetic peptide was analyzed using mass spectrometry and the resulting isotopic envelope was used to determine r1 and r2 from equations 3 and 4. This peptide corresponds to that noted in blue in Figure 1A (inset). (B) The same synthetic peptide was incubated with 125 ng GST-LSD1 in demethylase buffer for two hours at 37°C. Samples were then subjected to MassSQUIRM analysis. A mixed population of overlapping peaks represents three different methylation states as seen in Figure 1C (inset). Peptides initially mono-methylated occur at a mass 2 Da heavier than di-methylated peptides due to the addition of one heavy methyl group during reductive methylation. Similarly, peptides initially unmodified will occur 4 Da heavier than di-methylated peptides due to the addition of two heavy methyl groups. Areas under the monoisotopic peaks were noted as A1, A2 and A3. These values were used to determine equations 5–7, which take into account the isotopic overlap noted in (A). Open circles indicate light methylation while closed circles indicate heavy methylation.