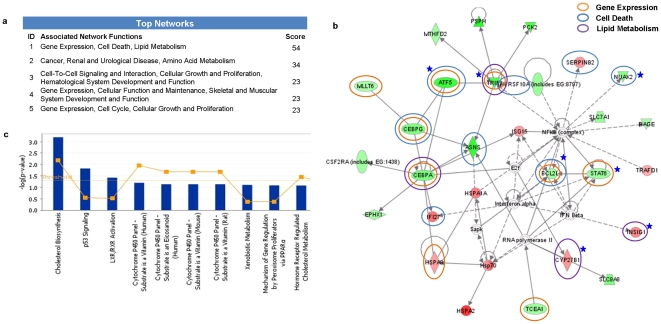
Figure 2. Ingenuity Pathways Analysis (IPA) summary.
To investigate possible interactions of differently regulated genes, datasets representing 273 genes with altered expression profile obtained from the illumina microarray were imported into the Ingenuity Pathway Analysis Tool and the following data is illustrated: a) The list of top five networks with their respective scores obtained from IPA b) Most highly rated network in IPA analysis The network representation of the most highly rated network (Gene Expression, Cell Death, Lipid Metabolism). The genes that are shaded were determined to be significant from the statistical analysis. The genes shaded red are upregulated and those that are green are downregulated. The intensity of the shading shows to what degree each gene was up or downregulated. A solid line represents a direct interaction between the two gene products and a dotted line means there is an indirect interaction. The genes marked with blue asterisk have been validated by real time PCR. Genes associated with Gene expression, Cell Death and Lipid Metabolism are circled with orange, blue and purple colours, respectively. c) Toxicology pathway list in IPA analysis. The x-axis represents the top toxicology functions as calculated by IPA based on differentially expressed genes are highlighted and the y-axis represents the ratio of number of genes from the dataset that map to the pathway and the number of all known genes ascribed to the pathway. The yellow line represents the threshold of p value < 0.05 as calculated by Fischer's test.