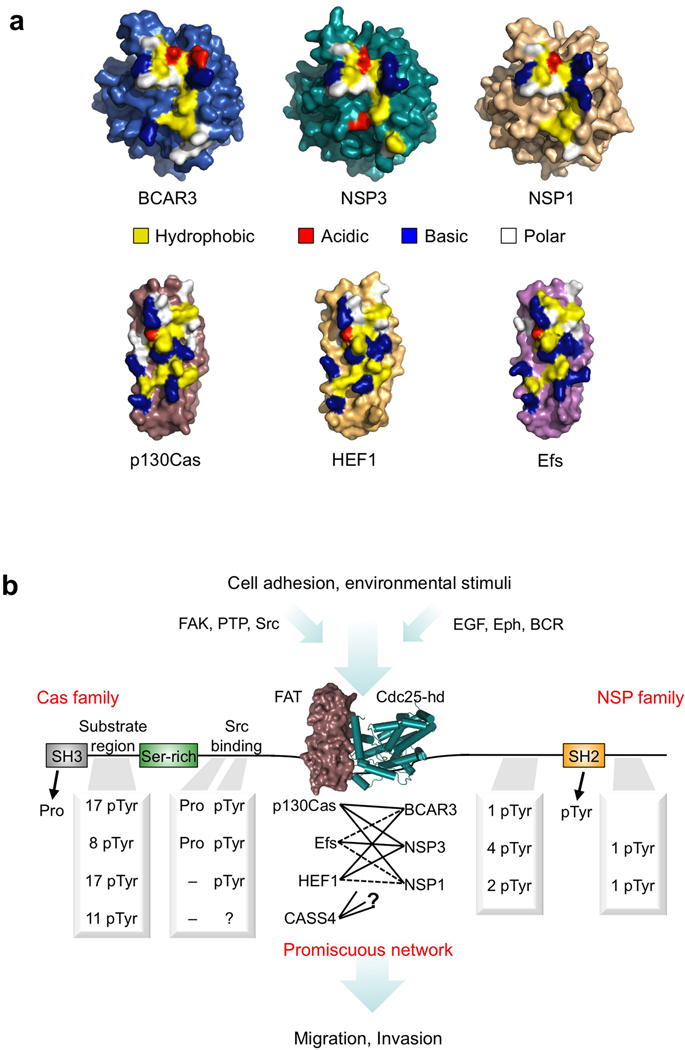
Figure 5.
NSP–Cas modules: Class-specific yet promiscuous signaling nodes. (a) Surface representations of the BCAR3, NSP3 and p130Cas structures solved in this work, as well as models of NSP1, HEF1 and Efs. Key residues involved in the NSP–Cas interaction are colored in yellow, blue, red or white indicating hydrophobic, basic, acidic or polar amino acids respectively. (b) Schematic representation of NSP–Cas signaling nodes. Association between the C-terminal domains of Cas and NSP family members forms a crucial junction downstream of adhesion and environmental stimuli, bringing together pairs of multifaceted signaling proteins that regulate cellular migration and invasion. The C-terminal association is of high-affinity but promiscuous between family members, and allows different combinations of docking sites and signaling outputs based upon the specific NSP–Cas module formed. Phosphorylation sites (pTyr) are annotated based on (www.phosphosite.org), except for Efs, which is based on ref 10. Proline motifs (Pro) for Src family kinase binding were identified manually based upon the sequence of each Cas family protein. (Abbreviations used as follows: PTP, Protein tyrosine phosphatases; EGF, epidermal growth factor; Eph, Ephrin receptors; BCR, B-cell receptor)