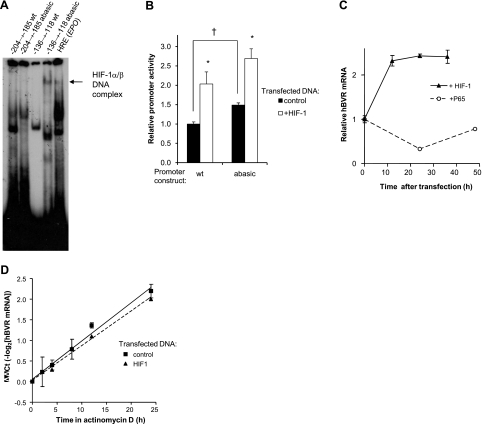
Figure 8.
Hypoxia activates hBVR transcription. A) Electrophoretic mobility shift assay. HEK293A cells were transfected with HIF-1α and ARNT expression plasmids; at 24 h after transfection, cells were transferred to 1% O2 for 24 h. The cells were lysed in the hypoxic chamber, and the lysates were immediately frozen in dry ice-ethanol. Aliquots of the lysate were assayed for binding to labeled oligonucleotides (located between nts −204 and −185, and −136 and −118), each containing a candidate HIF-binding site. An abasic site was introduced into the latter sequence by replacing G−131 in the complementary strand with dU, and treating the duplex with uracil glycosylase. Resolution and detection of binding products was as in Fig. 5A. Arrow marks a band observed only in hypoxia extracts. B) Abasic sites in the hBVR promoter enhance its activity. A 107-bp segment of hBVR promoter (nt −215 to −109), including the two most start-site proximal HREs, was assembled from 4 oligonucleotides and cloned into the pGL3-promoter plasmid (Materials and Methods). Two versions of the plasmid were built, a wild-type, dG-containing form, and one with dU substitutions in the putative HRE elements. Cells were cotransfected with the assembled construct, together with the pCMV-βgal plasmid and either empty pcDNA3 vector or the pcDNA3-HIFα and pcDNA3-ARNT plasmids. At 18 h after transfection, cells were lysed and assayed for luciferase and β-galactosidase activities. *P<0.01 vs. control; †P<0.001. C) Overexpression of HIF-1 increases hBVR mRNA. Cells were transfected with both pcDNA3-HIFα and pcDNA3-ARNT, and samples were taken at 12-h intervals for RNA isolation. cDNA was prepared, and the hBVR mRNA level was measured by quantitative PCR, normalized on 18S rRNA. Similarly, cells transfected with NF-κB P65 were assayed for hBVR mRNA expression. D) HIF-1 overexpression does not affect hBVR mRNA stability. Cells transfected with pcDNA3-HIFα and pcDNA3-ARNT or with empty vector were treated with 2.5 μg/ml actinomycin D, harvested at the indicated times, and hBVR mRNA was measured by quantitative PCR, as in C. mRNA half-life was calculated as the reciprocal of the slope of the linear regression.