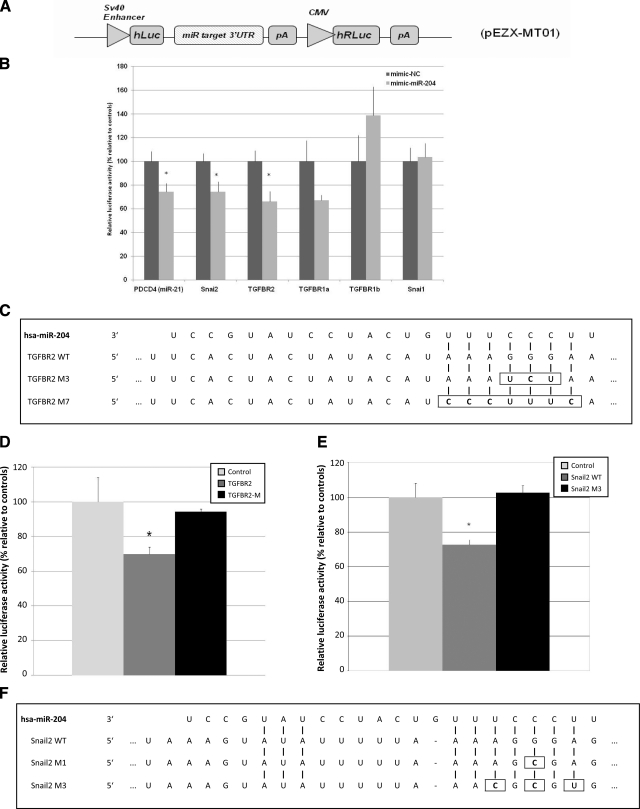
Figure 10.
A) Schematic diagram of the luciferase reporter vector pEZX-MT101. SV40, simian virus 40. B) Results of the luciferase assay demonstrating direct binding between miR-204 and its potential target mRNAs. Relative level of luciferase activity is measured by the ratio of firefly vs. Renilla luciferase activities normalized to mimic-NC (control mimic, a C. elegans miRNA that shares minimal sequence similarity to mammalian miRNAs). Data points are means ± se of 3 separate experiments. Candidate is considered a direct target if the magnitude of decrease in luciferase activity is equal to or greater than what is measured for miR-21 and its known target PDCD4 3′-UTR (positive control) (73). C) Alignments between hsa-miR-204 and its seed binding sequence in the 3′-UTR of TGF-βR2. In the M3 and M7 mutants of the pEZX-MT01 TGF-βR2-3′-UTR vector either 3-bp (M3) or 7-bp (M7) mutations were introduced within the seed binding sequence of the wild-type 3′-UTR by site-directed mutagenesis. D) Validation of TGF-βR2 as a direct target of miR-204 by site-directed mutagenesis. HEK293 cells were either transfected with a TGF-βR2 or a TGF-βR2 M7 or cotransfected with either the wild-type (TGF-βR2) or M7 version of the pEZX-MT01 TGF-βR2-3′-UTR vector with a miR-204 mimic, and the luciferase activity was normalized to vector only control (control, as 100%). E) Luciferase activities mediated by the 3′-UTR of the human SNAIL2 in the reporter construct pEZX-MT01 SNAIL2–3′-UTR. HEK293 cells in triplicate were treated as in D. F) Sequence alignments of the seed region of hsa-miR-204 with its target site in the 3′-UTR of human SNAIL2. WT, wild-type sequence of the target site; M1 and SNAIL2 M3, mutants of the wild-type target with 1- or 3-bp substitutions (enclosed by rectangles). Values are means ± se of 3 separate experiments. Values of P < 0.05 were considered significant; Student’s t test.