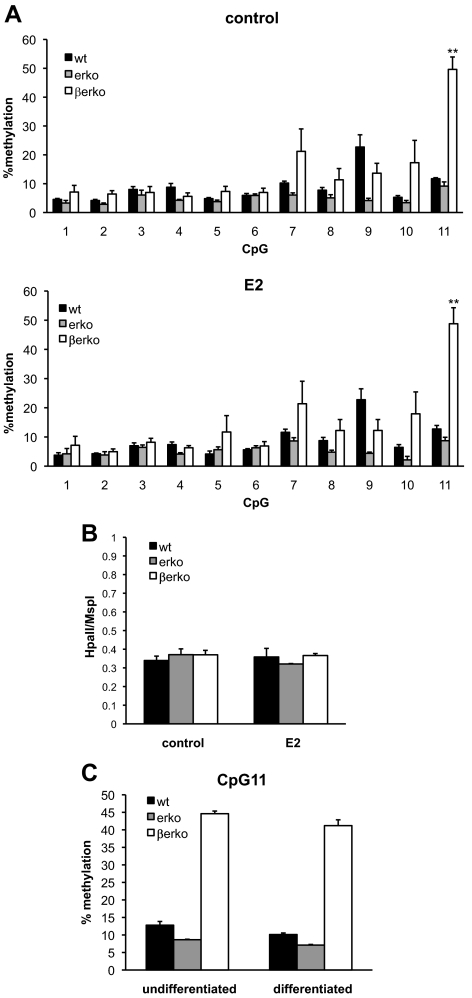
Fig. 2.
DNA methylation analyses in wt, erko, and βerko MEFs. A, Methylation analysis of CpG island 1 of the Glut4 promoter in the absence (upper panel) and presence (lower panel) of E2. Genomic DNA was isolated from wt, erko, and βerko MEFs, treated with 10 nm E2 for 4 d. DNA was bisulfite treated and sequenced by pyrosequencing. Data are represented as mean ± sd of three independent experiments. **, P < 0.01 βerko compared with wt and erko. B, Analysis of global DNA methylation using LUMA. MEFs derived from wt, erko, and βerko mice were treated with 10 nm E2 for 4 d, and genomic DNA was analyzed by digest with the restriction enzymes HpaII and MspI followed by pyrosequencing of the resulting fragments. Data are represented as mean + sd of three independent experiments. C, Methylation analysis after differentiation. MEFs were differentiated for 8 d, and genomic DNA was analyzed by bisulfite treatment and pyrosequencing. Shown is percent methylation at CpG11. Data are represented as mean ± sd.