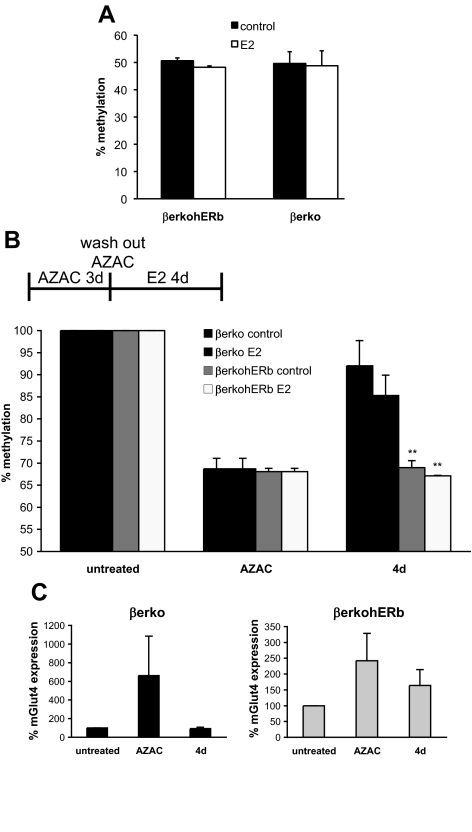
Fig. 3.
DNA methylation analysis of βerko MEFs after reintroduction of ERβ. A, Methylation analysis of the CpG11 in Glut4 promoter in the absence and presence of E2. Genomic DNA was isolated from βerko and βerko hERβ MEFs that were treated with 10 nm E2 for 4 d. DNA was bisulfite treated and sequenced by pyrosequencing. Data are represented as mean ± sd. B, Methylation analysis of CpG11 after DNA methyltransferase inhibition by 5-AZA-dC treatment. βerko and βerko hERβ MEFs were treated with 5-AZA-dC for 3 d, 5-AZA-dC was washed out, and cells were allowed to grow for another 4 d in the absence or presence of E2. Genomic DNA was then isolated, bisulfited treated, and analyzed by pyrosequencing. Methylation before 5-AZA-dC treatment was set to 100%. Data are represented as mean ± sd. **, P < 0.01, βerko hERβ compared with βerko. C, Glut4 expression after DNA methyltransferase inhibition by 5-AZA-dC treatment. βerko and βerko hERβ MEFs were differentiated and subsequently treated with 5-AZA-dC for 3 d, 5-AZA-dC was washed out, and cells were allowed to grow for another 4 d. RNA was extracted and analyzed after differentiation (untreated), after 5-AZA-dC treatment (AZAC), and after 4-d wash out (4 d) using real-time PCR. Data are normalized to GAPDH expression and Glut4 expression after differentiation was set to 100%. Data are represented as mean ± sd.