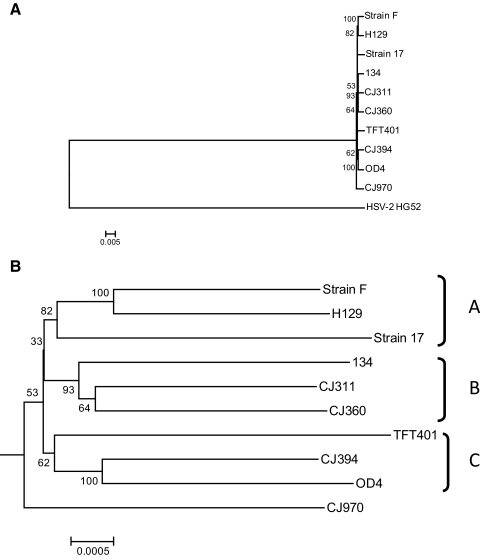
Figure 3.
Whole genome phylogenetic analysis of multiple HSV-1 viral strains. The HSV-1 genomes analyzed include the seven ocular isolates sequenced in this work, as well as the previously published genomes of strains 17, F, and H129. (A) Consensus bootstrap neighbor-joining tree using HSV-2 strain HG52 as an outgroup. (B) Expansion of the HSV-1 specific node from the neighbor-joining tree in (A). The genomes were aligned with ClustalW and then consensus bootstrap (1000 replicates) neighbor-joining trees, using the Tamura-Nei algorithm, were generated with the Mega4 package. The phylogenetic distance is located at the bottom of each tree.