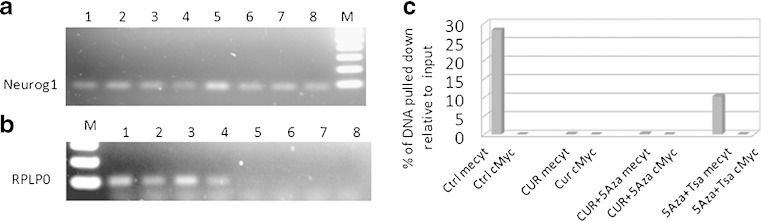
Fig. 3.
MeDIP analysis of Neurog1 methylation. Eight-microgram genomic DNAs extracted from control (Ctrl), curcumin (CUR), curcumin/5-Aza-2-deoxycytidine (CUR/5-Aza), 5-Aza/Trichostatin A (5-Aza/TSA, TSA was added 20 h before harvested) were sonicated, denatured, and subjected to DNA IP with anti-methyl cytosine (mecyt) antibody. a, b Semi-quantitative PCR comparing the immunoprecipitated DNA with its inputs; lanes 1–4 input DNA of (1 = Ctrl, 2 = CUR, 3 = CUR/5-Aza, 4 = 5-Aza/TSA); lanes 5–8 Mecyt-IP DNA of (5 = Ctrl; 6 = CUR; 7 = CUR/5-Aza; 8 = 5-Aza/TSA). M marker. Primers used are a Neurog1 and b RPLP0. c qPCR was performed to quantify the IP DNA with its inputs. The inputs were diluted from 2×, 4×, 8×, 16×, and 32×, based on the standard curve of ΔCT value, and the relative amount of IP DNA was calculated. The cMyc antibody was considered as non-specific binding control