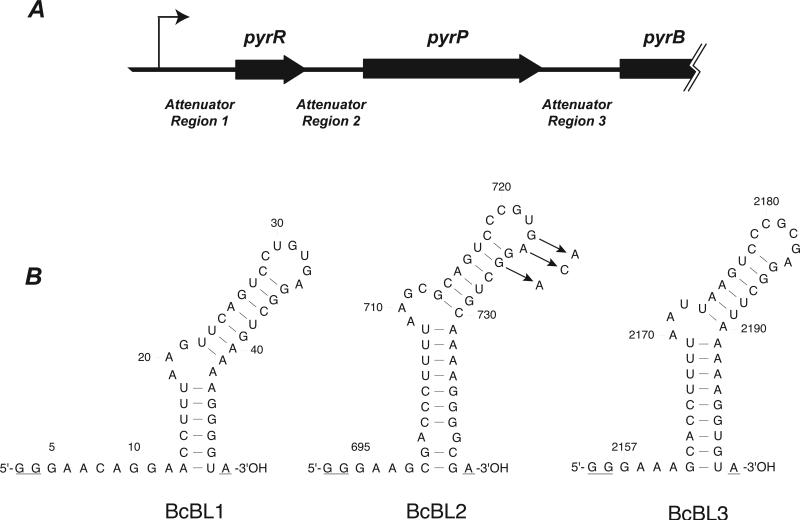
Figure 1.
(A) Map of the 5’-end of the B. caldolyticus pyr operon. The thin bent arrow represents the transcriptional start site; open reading frames are represented as thick arrows; the untranslated regions containing the three attenuator regions are shown as lines of medium thickness. (B) Sequence of the three pyr mRNA species (binding loops) bound by PyrR that were examined in this study. The BcBL1, BcBL2 and BcBL3 sequences were derived from portions of the DNA sequence of Attenuator Regions 1, 2 and 3, respectively, shown in A. Numbers refer to the nucleotide number in the B. caldolyticus pyr transcript with +1 as the transcriptional start site [13]. The secondary structures were predicted by MFOLD version 3.1 (http://www.bioinfo.rpi.edu/applications/mfold) [33]. Three nucleotides in each binding loop that are not part of the wild type pyr mRNA sequence are underlined: The two first G residues in each transcript are specified by the T7 promoter and the terminal A residue is added by Taq polymerase when used for preparation of templates for in vitro transcription by T7 polymerase. Arrows indicate three single-base substitution RNA variants in BcBL2 examined in this study.