This report describes the crystallization and X-ray diffraction analysis of two different allelic variants of the AvrM effector from flax rust.
Keywords: AvrM, fungal effectors, plant disease resistance, single-wavelength anomalous dispersion
Abstract
The flax rust effector AvrM is a secreted protein of unknown fold that is recognized by the M resistance protein in flax. In order to investigate the structural basis of the AvrM–M interaction and possible virulence-associated functions of AvrM, the C-terminal domains of two different AvrM variants (AvrM-A and avrM) were crystallized. Crystals of native AvrM-A were obtained using pentaerythritol ethoxylate (15/4 EO/OH) as a precipitant and diffracted X-rays to 2.9 Å resolution. Selenomethionine-derivative crystals of similar quality were obtained using PEG 1500 as a precipitant. Both the native and selenomethionine-labelled AvrM-A crystals had symmetry of space group C2221 with eight molecules in the asymmetric unit. Crystals of avrM had symmetry of space group P212121 and diffracted X-rays to 2.7 Å resolution. Initial AvrM-A phases were calculated using the single-wavelength anomalous dispersion (SAD) method and a partial model was built. Phases for avrM were obtained by molecular replacement using the partial AvrM-A model.
1. Introduction
Many plant pathogens deliver effector molecules into the plant cell, where they promote virulence, often by interfering with basal immune signalling. However, plants have evolved specific immune receptors known as resistance (R) proteins that recognize these effectors through direct or indirect mechanisms, resulting in effector-triggered immunity (ETI; Chisholm et al., 2006 ▶; Jones & Dangl, 2006 ▶; Rafiqi et al., 2009 ▶; Dodds & Rathjen, 2010 ▶). ETI is characterized by localized cell death, termed the hypersensitive response (HR), at the site of infection that limits the spread of the pathogen. The effectors that are detected by the R proteins are often referred to as avirulence (Avr) proteins because their presence enables the plant to engage an effective defence response, resulting in the plants being immune to pathogen infection.
The majority of R proteins are structurally conserved and have a central nucleotide-binding (NB) domain, a C-terminal leucine-rich-repeat (LRR) domain and either a coiled-coil (CC) domain or a Toll-interleukin-1 receptor (TIR) domain at the N-terminus (Martin et al., 2003 ▶). The LRR domain appears to be involved in both recognition of the effector proteins and negative regulation of the HR (Ellis et al., 1999 ▶; Dodds et al., 2001 ▶; Rairdan & Moffett, 2006 ▶; Padmanabhan et al., 2009 ▶), while the NB and TIR domains are required for activation of the R proteins and initiation of defence signalling (Frost et al., 2004 ▶; Lukasik & Takken, 2009 ▶; Swiderski et al., 2009 ▶; Krasileva et al., 2010 ▶; Bernoux et al., 2011 ▶; Williams et al., 2011 ▶).
Plant pathogen effectors have few common features and most of them lack characterized homologues, making it difficult to infer any molecular function from their sequence alone. Many bacterial effectors are secreted directly into the host cytosol through the type 3 secretion system and have been shown to interfere with basal immune responses (Hann et al., 2010 ▶). Fungal and oomycete effectors are also delivered into the host cell (Panstruga & Dodds, 2009 ▶; Tyler, 2009 ▶; Rafiqi et al., 2010 ▶; Kale et al., 2010 ▶), but data on their molecular function and virulence targets are scarce.
The interaction between flax (Linum usitatissimum) and the fungal rust pathogen Melampsora lini is a powerful model system for studying ETI. Several R genes and corresponding flax rust effector genes have been identified and some of the effectors, including AvrL567 and AvrM, encode secreted proteins that are recognized inside the plant cell by R proteins that belong to the TIR–NB–LRR class (Dodds et al., 2004 ▶; Catanzariti et al., 2006 ▶; Wang et al., 2007 ▶). The AvrM effector is recognized by the M resistance protein in flax and a direct interaction between AvrM and M has been demonstrated (Catanzariti et al., 2010 ▶). In the rust CH5 strain six different variants (AvrM-A, AvrM-B, AvrM-C, AvrM-D, AvrM-E and avrM) have been identified, but only four of these proteins (AvrM-A, AvrM-B, AvrM-C and AvrM-D) are recognized by M and induce HR. AvrM protein sequences are not related to any other sequences available in the current databases and their virulence functions and cellular targets are unknown. Recently, it has been shown that the C-terminal region of AvrM-A forms a structured domain which is required for recognition by M in flax and for direct interaction with M in yeast two-hybrid experiments. Biophysical characterization revealed that the full-length protein has a α-helical fold and is a stable dimer in solution (Catanzariti et al., 2010 ▶).
In order to understand the structural basis of the AvrM–M interaction and to provide insights into possible virulence-associated functions of AvrM, we have embarked on determination of its three-dimensional structure. Here, we report the crystallization and X-ray diffraction analysis of the C-terminal domain of AvrM-A (residues 103–343) and avrM (residues 46–280). The protein avrM encoded by the ‘virulence allele’ is not recognized by M and shares 94% sequence identity with AvrM-A in the C-terminal domain.
2. Materials and methods
2.1. Protein production and purification
Gene fragments encoding residues 103–343 of AvrM-A and residues 46–280 of avrM (here termed simply ‘AvrM-A’ and ‘avrM’, respectively) were amplified by PCR using the forward primers 5′-TACTTCCAATCCAATGCGCAACCAGAATTTGACAGAGGATTCC-3′ and 5′-TACTTCCAATCCAATGCGGTTGAAGGGATCCCTCAACCAG-3′, respectively, and the same reverse primer 5′-TTATCCACTTCCAATGTTACATGTCGGAGATTTCAATATCTTGTTC-3′. The PCR products were inserted into the pMCSG7 expression vector using ligation-independent cloning (Stols et al., 2002 ▶). The integrity of the resulting constructs was verified by sequencing. Both proteins were expressed in Escherichia coli BL21 (DE3) cells using autoinduction medium (Studier, 2005 ▶) containing 100 µg ml−1 ampicillin. The cells were grown at 310 K to mid-exponential phase (OD600nm of approximately 0.6–0.8). The temperature was then reduced to 293 K and the cultures were grown for approximately 16 h. The cells were harvested by centrifugation at 5000g and stored at 193 K. Selenomethionine-labelled AvrM-A was produced in E. coli methionine-auxotroph B834 (DE3) cells (Novagen). The cells were grown at 310 K in M9 minimal medium supplemented with 0.2 mM selenomethionine (SeMet) until an OD600nm of 0.6 was reached. The temperature was then reduced to 293 K and protein expression was induced after 1 h by the addition of isopropyl β-d-1-thiogalactoside to a final concentration of 1 mM. Growth was continued for a further 16 h at 293 K. The cells were resuspended in 5 ml pre-chilled lysis buffer (50 mM HEPES pH 8.0, 500 mM NaCl) per gram of cells and lysed using a digitial sonifier (Branson). The cell debris was removed by centrifugation at 15 000g for 30 min at 277 K and the resulting supernatant was applied onto a 5 ml HisTrap FF column (GE Healthcare). The column was washed with 20 column volumes of lysis buffer containing 30 mM imidazole and the protein was eluted using a linear gradient of imidazole from 30 to 250 mM over 20 column volumes. The fractions containing AvrM-A and avrM were pooled and the N-terminal His6 tag was removed by overnight treatment with His6-tagged tobacco etch virus (TEV) protease (100 µl at 8 mg ml−1 per 20 ml of sample) at 277 K in TEV cleavage buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mM DTT, 0.5 mM EDTA). Prior to cleavage, the protein was buffer-exchanged by concentrating the pooled sample to 2 ml using Amicon ultracentrifugal devices (10 000 Da molecular-weight cutoff; Millipore), followed by subsequent dilution to 20 ml in TEV cleavage buffer. TEV-cleaved AvrM-A and avrM were purified from the TEV reaction mixture using the same IMAC strategy as described above. Column-flowthrough fractions containing cleaved protein were pooled together and concentrated. AvrM-A and avrM were further purified by gel filtration on a Superdex 200 HiLoad 26/60 column (GE Healthcare) pre-equilibrated with 10 mM HEPES pH 7.4, 150 mM NaCl. The same protocol was used for the purification of the SeMet-labelled AvrM-A, but 1 mM DTT was included in all buffers. The peak fractions of AvrM-A and avrM were pooled and concentrated to final concentrations of 20–90 and 28 mg ml−1, respectively, in gel-filtration buffer. The concentrated protein samples were flash-cooled as 25 µl aliquots in liquid nitrogen and stored at 193 K. The protein concentration was determined by measuring the absorbance at 280 nm using an extinction coefficient of 13 410 M −1 cm−1 for both AvrM-A and avrM (Expasy ProtParam tool; http://ca.expasy.org/cgi-bin/protparam).
2.2. Crystallization and X-ray diffraction analysis
Initial screening of crystallization conditions was performed in 96-well plates (LabTech) at 293 K using the hanging-drop vapour-diffusion technique. Several commercial screens were employed, including Index, PEG/Ion and PEGRx (Hampton Research), Pact Premier and JCSG+ (Qiagen), Synergy and Axygen (Jena Biosciences) and ProPlex (Molecular Dimensions). 200 nl drops consisting of 100 nl protein solution and 100 nl reservoir solution were prepared on hanging-drop seals (TTP4150-5100 sourced from Millennium Science, Australia) using a Mosquito robot (TTP LabTech, UK) and equilibrated against 100 µl reservoir solution. The drops were monitored and imaged using the Rock Imager system (Formulatrix, USA).
Hits from the initial crystallization screens were optimized by varying the protein concentration, the precipitant concentration, the pH and the size of the drop. For optimization using streak-seeding, a cat whisker was passed through a drop containing native crystals and subsequently passed through 4–6 fresh crystallization drops in a 24-well sitting-drop plate (Cryschem, Hampton Research).
For data collection under cryogenic conditions (100 K), native AvrM-A crystals were flash-cooled by plunging them directly into a liquid-nitrogen bath, while SeMet-labelled AvrM-A crystals were transferred into mother-liquor solution containing 20% ethylene glycol prior to freezing. The avrM crystals were soaked in Paratone-N (Hampton Research) before flash-cooling in liquid nitrogen.
Native data sets from single crystals of both AvrM-A and avrM were collected on the Australian Synchrotron MX2 beamline at wavelengths of 0.978508 and 0.953940 Å, respectively, using an ADSC Quantum 315r CCD detector. For AvrM-A 360 images were collected, the crystal-to-detector distance was set to 350 mm and the oscillation range was 0.5°. For avrM, the crystal-to-detector distance was set to 400 mm, the oscillation range was 1.0° and 180 images were collected. Data for the SeMet derivative of AvrM-A were collected on the MX1 beamline of the Australian Synchrotron using an ADSC Quantum 210r CCD detector. A fluorescence scan was taken around the selenium absorption edge and diffraction data were collected at the absorption peak (0.979425 Å). A total of 720 images were collected with an oscillation range of 0.5° and a crystal-to-detector distance of 300 mm.
Data were indexed and integrated using either MOSFLM (Leslie, 1992 ▶) or XDS (Kabsch, 2010 ▶) and scaled with SCALA within the CCP4 suite (Winn et al., 2011 ▶). SAD phasing was carried out using the AutoSol automated search routine from the PHENIX program suite (Adams et al., 2010 ▶).
3. Results and discussion
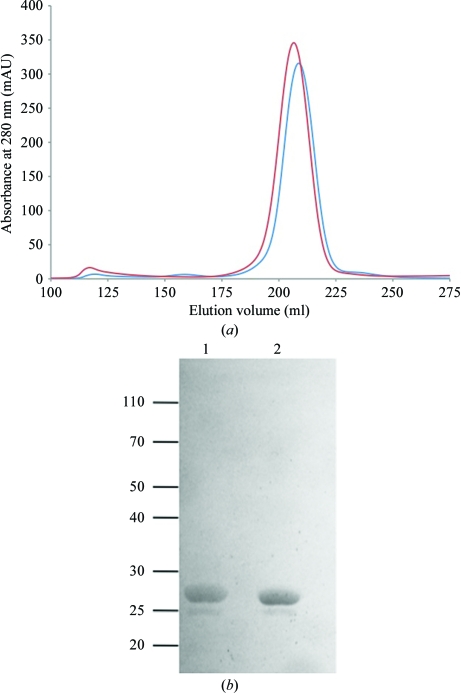
The C-terminal domains of AvrM-A and avrM were purified to >95% homogeneity as judged by SDS–PAGE (Fig. 1 ▶ b), with yields of 15 and 10 mg protein per litre of culture, respectively. The gel-filtration profiles of both proteins (Fig. 1 ▶ a) are consistent with monodisperse protein with a molecular weight corresponding to a dimer, which is consistent with previous observations for full-length AvrM-A (Catanzariti et al., 2010 ▶). Initial crystallization screens of purified AvrM-A were performed using drops of 200 nl volume in 96-well microplates and different protein concentrations were trialled (20, 45 and 90 mg ml−1). AvrM-A crystals were observed after 3–5 d at 20 mg ml−1 in formulations No. 5 (0.1 M SPG pH 8, 25% PEG 1500), No. 16 (0.1 M MIB pH 6, 25% PEG 1500) and No. 38 (0.1 MMT pH 6, 25% PEG 1500) of Pact Premier and at 90 mg ml−1 in formulation No. 57 [50 mM bis-tris pH 6.5, 50 mM ammonium sulfate, 30% pentaerythritol ethoxylate (15/4 EO/OH)] of the Index screen. The crystals from the latter condition (Fig. 2 ▶ a) attained final dimensions of 300 × 200 × 100 µm after three weeks and a complete data set to 2.9 Å resolution was collected at the Australian Synchrotron from a single crystal grown from this condition. Analysis of the diffraction data using POINTLESS (Evans, 2006 ▶) identified an orthorhombic lattice and C2221 as the most likely space group based on systematic absences. The unit-cell parameters for the AvrM-A crystals were a = 116.8, b = 131.2, c = 280.3 Å. The crystals have a Matthews coefficient (Matthews, 1968 ▶) of 2.41 Å3 Da−1 and a solvent content of 49% assuming eight protein molecules per asymmetric unit.
Figure 1.
Analysis of AvrM-A and avrM by gel filtration and SDS–PAGE. (a) Elution profiles of AvrM-A (red) and AvrM (blue) on a Superdex 200 26/60 column. (b) SDS–PAGE gel of the peak fractions from the chromatogram in (a). Lanes 1 and 2 correspond to AvrM-A and avrM, respectively. Approximately 5 µg protein from the peak fractions was loaded onto the gel.
Figure 2.
Crystals of AvrM-A and avrM. (a) Native crystals of AvrM-A grown using pentaerythritol ethoxylate (15/4 EO/OH) with dimensions of 300 × 200 × 100 µm. (b) Similar-sized SeMet-derivative crystals of AvrM-A obtained using PEG 1500 as precipitant. (c) Crystals of avrM grown using sodium citrate. The longest edge corresponds to approximately 400 µm.
As there is no homologue of AvrM of known structure, the phase problem had to be solved experimentally. We therefore attempted to obtain crystals of an SeMet-labelled derivative of AvrM-A. Initial optimization attempts varying the precipitant concentration, protein concentration, pH and drop size failed to produce both native and SeMet-derivatized crystals using the PEG 1500-containing conditions. SeMet-derivatized and native crystals could be produced using pentaerythritol ethoxylate (15/4 EO/OH), but high nucleation yielded small crystals that did not diffract X-rays beyond 5 Å resolution. Attempts to lower the nucleation rate by using additives (Additive Screen HT, Hampton Research), different drop ratios and crystallization under oil (Chayen, 1997 ▶) failed to produce larger crystals. However, when the pentaerythritol ethoxylate (15/4 EO/OH) was replaced by PEG 1500 the rate of nucleation was reduced and the size of the crystals was greatly improved. SeMet-derivatized crystals diffracting X-rays to 3.0–3.5 Å resolution were obtained in 20–22% PEG 1500, 50 mM bis-tris pH 5.9–6.2, 50 mM ammonium sulfate using a protein concentration of 45 mg ml−1 (Fig. 2 ▶ b). Similar SeMet-derivatized crystals were also obtained by streak-seeding of native crystals in 0.1 M MMT buffer pH 5.6–6.0, 25% PEG 1500.
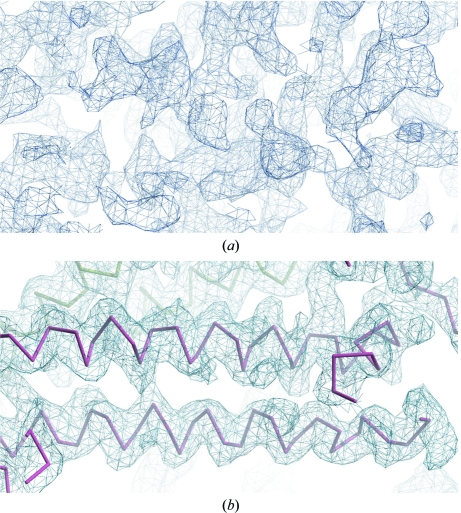
Phase information was obtained from an Se-SAD data set at 3.5 Å resolution collected from a single crystal grown from seeds in 0.1 M MMT buffer pH 5.8, 25% PEG 1500. The crystal had symmetry of the same space group as the native crystals and a total of 22 selenium sites were identified in the asymmetric unit. The initial phases calculated using AutoSol within the PHENIX suite (Adams et al., 2010 ▶; Fig. 3 ▶ a) had a figure of merit (FOM) of 0.29. The phases were substantially improved during density modification to give a final FOM of 0.75 and the resulting electron density (Fig. 3 ▶ b) was of sufficient quality to allow tracing of about 75% of the main-chain atoms of all eight copies in the asymmetric unit. Manual model building and refinement of the structure is currently in progress.
Figure 3.
(a) Experimental electron-density map of one section of the asymmetric unit after initial phasing using AutoSol within PHENIX. (b) The same section after density modification, with the partially traced main chains shown in red and green (symmetry-related molecule). Both maps were produced in Coot (Emsley & Cowtan, 2004 ▶) and contoured at 1.0σ.
In parallel to reproducing and optimizing crystals of AvrM-A, crystallization screens for avrM were also set up using a concentration of 28 mg ml−1. Rod-shaped crystals were observed after 12–24 h in 0.1 M HEPES pH 7.5, 1.6 M sodium citrate tribasic dihydrate (Index formulation No. 20) and in 1.9 M sodium malonate pH 6 (Synergy formulation No. 85). Optimized crystals grew within 5 d in 0.1 M MES pH 6.5, 1.3 M sodium citrate using a protein concentration of 10 mg ml−1 (Fig. 2 ▶ c). Data sets to 2.7 Å resolution were collected and analysis of the systematic absences suggested P212121 as the most likely space group. Data-collection statistics are given in Table 1 ▶. Molecular replacement was performed using Phaser (McCoy et al., 2007 ▶) with a partial model of the AvrM-A monomer as the search model. Phasing statistics are given in Table 2 ▶. One solution consisting of four monomers was found in space group P212121 with a final translation-function Z score of 16.5 and a log-likelihood gain of 787. We now expect that the three-dimensional structures of both AvrM variants can be determined using the data collected.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| AvrM-A | SeMet AvrM-A | avrM | |
|---|---|---|---|
| Space group | C2221 | C2221 | P212121 |
| Unit-cell parameters (Å) | |||
| a | 116.8 | 116.4 | 88.5 |
| b | 131.2 | 133.7 | 125.7 |
| c | 280.3 | 281.5 | 128.9 |
| Molecules per asymmetric unit | 8 | 8 | 4 |
| Resolution range (Å) | 30–3.0 (3.16–3.00) | 93.8–3.5 (3.69–3.50) | 19.9–2.7 (2.85–2.70) |
| No. of unique observations | 43446 | 28160 | 40138 |
| Completeness (%) | 99.9 (99.9) | 100.0 (100.0) | 99.7 (100.0) |
| Multiplicity | 6.0 (6.1) | 14.9 (15.0) | 7.3 (7.4) |
| Rmerge† (%) | 10.2 (85.7) | 19.0 (74.0) | 8.5 (76.6) |
| Average I/σ(I) | 13.1 (2.3) | 13.1 (4.4) | 15.0 (2.7) |
R
merge = 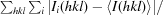
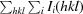 , where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I
hkl〉 is the mean intensity of that reflection. Calculated for I > −3σ(I).
, where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I
hkl〉 is the mean intensity of that reflection. Calculated for I > −3σ(I).
Table 2. Phasing statistics.
| AutoSol | |
| No. of sites (found/all) | 24/32 |
| HySS† CC‡ | 0.33 |
| SKEW§ | 0.09 |
| CORRRMS¶ | 0.75 |
| FOM†† | 0.29 |
| Estimated map CC | 0.28 ± 0.34 |
| After density modification | |
| R factor | 0.29 |
| FOM after density modification | 0.75 |
Hybrid Substructure Search.
Correlation coefficient.
Skew of the electron density in the map.
The correlation of local r.m.s. density.
Figure of merit.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., Zhang, X., Ellis, J. G., Kobe, B. & Dodds, P. N. (2011). Cell Host Microbe, 9, 200–211. [DOI] [PMC free article] [PubMed]
- Catanzariti, A. M., Dodds, P. N., Lawrence, G. J., Ayliffe, M. A. & Ellis, J. G. (2006). Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed]
- Catanzariti, A. M., Dodds, P. N., Ve, T., Kobe, B., Ellis, J. G. & Staskawicz, B. J. (2010). Mol. Plant Microbe Interact. 23, 49–57. [DOI] [PMC free article] [PubMed]
- Chayen, N. E. (1997). Structure, 5, 1269–1274. [DOI] [PubMed]
- Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. (2006). Cell, 124, 803–814. [DOI] [PubMed]
- Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Ayliffe, M. A. & Ellis, J. G. (2004). Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed]
- Dodds, P. N., Lawrence, G. J. & Ellis, J. G. (2001). Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed]
- Dodds, P. N. & Rathjen, J. P. (2010). Nature Rev. Genet. 11, 539–548. [DOI] [PubMed]
- Ellis, J. G., Lawrence, G. J., Luck, J. E. & Dodds, P. N. (1999). Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Frost, D., Way, H., Howles, P., Luck, J., Manners, J., Hardham, A., Finnegan, J. & Ellis, J. (2004). Mol. Plant Microbe Interact. 17, 224–232. [DOI] [PubMed]
- Hann, D. R., Gimenez-Ibanez, S. & Rathjen, J. P. (2010). Curr. Opin. Plant Biol. 13, 388–393. [DOI] [PubMed]
- Jones, J. D. & Dangl, J. L. (2006). Nature (London), 444, 323–329. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kale, S. D., Gu, B., Capelluto, D. G., Dou, D., Feldman, E., Rumore, A., Arredondo, F. D., Hanlon, R., Fudal, I., Rouxel, T., Lawrence, C. B., Shan, W. & Tyler, B. M. (2010). Cell, 142, 284–295. [DOI] [PubMed]
- Krasileva, K. V., Dahlbeck, D. & Staskawicz, B. J. (2010). Plant Cell, 22, 2444–2458. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Lukasik, E. & Takken, F. L. (2009). Curr. Opin. Plant Biol. 12, 427–436. [DOI] [PubMed]
- Martin, G. B., Bogdanove, A. J. & Sessa, G. (2003). Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Padmanabhan, M., Cournoyer, P. & Dinesh-Kumar, S. P. (2009). Cell. Microbiol. 11, 191–198. [DOI] [PMC free article] [PubMed]
- Panstruga, R. & Dodds, P. N. (2009). Science, 324, 748–750. [DOI] [PMC free article] [PubMed]
- Rafiqi, M., Bernoux, M., Ellis, J. G. & Dodds, P. N. (2009). Semin. Cell Dev. Biol. 20, 1017–1024. [DOI] [PubMed]
- Rafiqi, M., Gan, P. H., Ravensdale, M., Lawrence, G. J., Ellis, J. G., Jones, D. A., Hardham, A. R. & Dodds, P. N. (2010). Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed]
- Rairdan, G. J. & Moffett, P. (2006). Plant Cell, 18, 2082–2093. [DOI] [PMC free article] [PubMed]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R. & Donnelly, M. I. (2002). Protein Expr. Purif. 25, 8–15. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Swiderski, M. R., Birker, D. & Jones, J. D. (2009). Mol. Plant Microbe Interact. 22, 157–165. [DOI] [PubMed]
- Tyler, B. M. (2009). Cell. Microbiol. 11, 13–20. [DOI] [PubMed]
- Wang, C. I., Guncar, G., Forwood, J. K., Teh, T., Catanzariti, A. M., Lawrence, G. J., Loughlin, F. E., Mackay, J. P., Schirra, H. J., Anderson, P. A., Ellis, J. G., Dodds, P. N. & Kobe, B. (2007). Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed]
- Williams, S. J., Sornaraj, P., deCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., Dodds, P. N. & Anderson, P. A. (2011). Mol. Plant Microbe Interact. 24, 897–906. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.