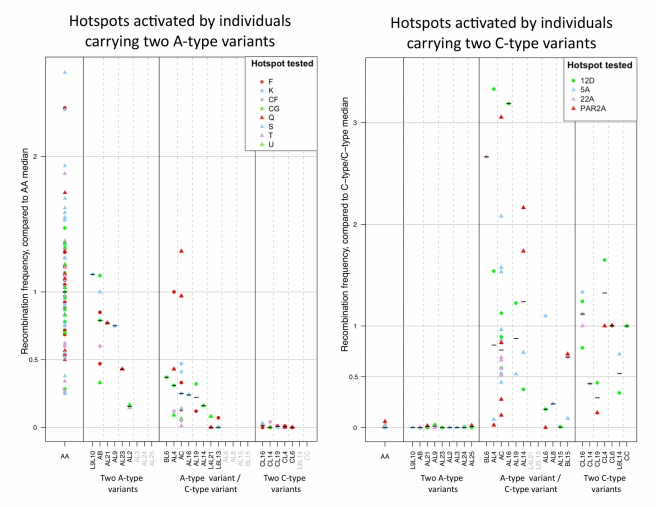
Figure 2. The effect of PRDM9 zinc finger variants on hotspot activity.
Each column presents males with the same genotype, grouped according to whether they carry two A-type variants (defined as variants predicted to bind the same 13-bp motif as A), two C-type variants (defined as variants predicted to bind the same 17-bp motif as C), or one A-type and one C-type variant. Within a column, each symbol denotes the recombination activity of a given hotspot for a given individual, with circles indicating hotspots that contain a perfect match to the 13-bp motif (for the left panel) or the 17-bp motif (for the right panel) within 1 kb of their center, and triangles indicating hotspots with no perfect matches. The median recombination frequency is shown as a black bar. As can be seen, there is no clear difference between the activity of hotspots with and without a perfect match to the motif. The recombination frequency is reported relative to the median of AA individuals (left panel) or that of C-type/C-type individuals (right panel). The data were obtained by sperm-typing from [18] (left panel) and [24] (right panel). The E and PAR2 hotspots from [18] were excluded from the analysis because they contain polymorphisms disrupting the central 13-bp motif [42], possibly confounding the effect of variation in PRDM9. The 12B hotspot from [24] was excluded because it was not active in typed C-type/C-type individuals.