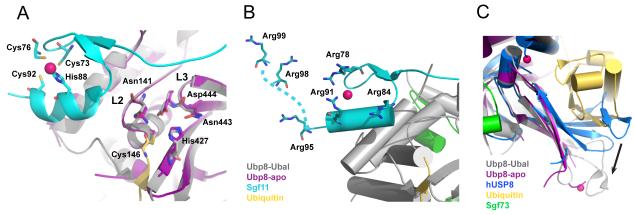
Figure 3. The activation of Ubp8 and substrate recognition of the DUBm.
A) The Sgf11 zinc finger binds adjacent to the Ubp8 active site and interacts with loops L2 and L3, which contain the predicted oxyanion hole residues N141, D444, and the catalytic residues, N443 and C146. Also shown is H427, the third residue in the catalytic triad. The active site residues adopt the same conformation in the absence (purple DUBm) and presence (grey DUBm) of ubiquitin (colors indicated in figure, zinc atoms are colored dark pink). B) Conserved arginine residues that form a basic patch on the Sgf11 zinc finger are indicated, including Arg98 and Arg99, which are disordered in the structure and have been modeled into the figure (indicated by the dashed line). C) The fingers region of Ubp8 adopts an open conformation in the presence (gray) and absence (magenta) of bound ubiquitin. By contrast, fingers region in the structure of human USP8 [36] occludes the ubiquitin-binding pocket, and must undergo a conformational change in order to bind ubiquitin, as indicated by the black arrow.