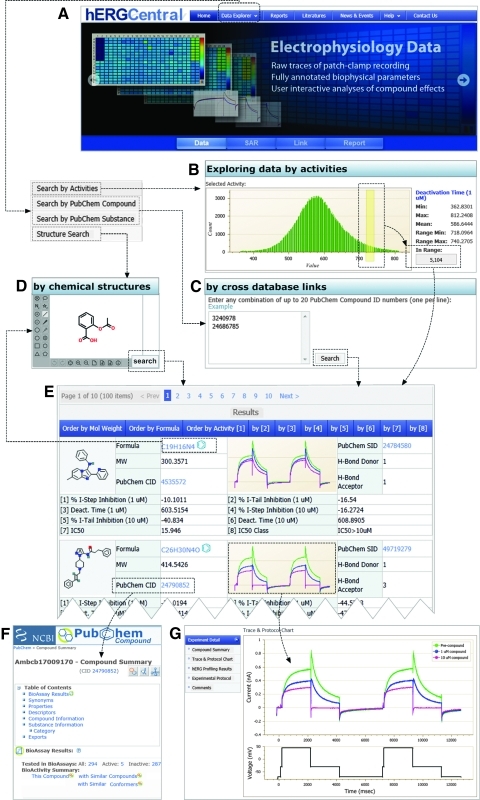
Fig. 1.
Overall outline of hERGCentral. (A) Home page of hERGCentral, on the top dropdown menu for three ways of data exploration can be accessed. (B) Exploring data by activities: large scale of hERG data can be explored interactively by various activities such as tail current inhibition. (C) Exploring data by cross database links: external database IDs (here, PubChem compounds and substances) can be used to browse data on hERGCentral. (D) Exploring data by chemical structures: molecular structures can be easily entered through the Web interface, which allows users to explore results with similar structures. (E) In either way of exploration, the results are presented in a tabular view, where various properties, activities, as well as thumbnails of raw current trace data are listed. By clicking any property or activity on the heading of this table, users can sort the results accordingly. (F) Live links to external public databases, for example, PubChem compound summary, are maintained. (G) Detailed results such as electrophysiological recording traces at a higher resolution, as well as voltage protocols can be accessed by clicking the current trace thumbnails. CID, compound ID; hERG, human Ether-à-go-go related gene; SID, substance ID; ID, identity.