Abstract
PA-824 is a promising drug candidate for the treatment of tuberculosis (TB). It is in phase II clinical trials as part of the first newly designed regimen containing multiple novel antituberculosis drugs (PA-824 in combination with moxifloxacin and pyrazinamide). However, given that the genes involved in resistance against PA-824 are not fully conserved in the Mycobacterium tuberculosis complex (MTBC), this regimen might not be equally effective against different MTBC genotypes. To investigate this question, we sequenced two PA-824 resistance genes (fgd1 [Rv0407] and ddn [Rv3547]) in 65 MTBC strains representing major phylogenetic lineages. The MICs of representative strains were determined using the modified proportion method in the Bactec MGIT 960 system. Our analysis revealed single-nucleotide polymorphisms in both genes that were specific either for several genotypes or for individual strains, yet none of these mutations significantly affected the PA-824 MICs (≤0.25 μg/ml). These results were supported by in silico modeling of the mutations identified in Fgd1. In contrast, “Mycobacterium canettii” strains displayed a higher MIC of 8 μg/ml. In conclusion, we found a large genetic diversity in PA-824 resistance genes that did not lead to elevated PA-824 MICs. In contrast, M. canettii strains had MICs that were above the plasma concentrations of PA-824 documented so far in clinical trials. As M. canettii is also intrinsically resistant against pyrazinamide, new regimens containing PA-824 and pyrazinamide might not be effective in treating M. canettii infections. This finding has implications for the design of multiple ongoing clinical trials.
INTRODUCTION
The Mycobacterium tuberculosis complex (MTBC) bacteria, the causative agents of tuberculosis (TB), are devastating pathogens. There is an urgent need for new drugs which are not only effective against multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB but also capable of shortening the length of treatment regimens, given that the prolonged treatment results in significant direct and opportunity costs to patients, who are often affected by poverty and structural violence in the first place (11). The latter criterion requires that the antibiotic is active against both replicating and dormant cells. PA-824, a novel nitroimidazole, satisfies all these conditions (12, 38). It has been evaluated extensively in animals and human subjects and has already entered phase II clinical trials as part of the first regimen (PA-824-moxifloxacin-pyrazinamide) that contains multiple new antituberculosis drugs (12, 20).
The aerobic killing mechanism of this prodrug involves the inhibition of cell wall mycolic acid biosynthesis, whereas the anaerobic mechanism works via respiratory poisoning, where PA-824 directly acts as an NO donor (24, 31, 37). This reaction is catalyzed by Ddn, a nitroreductase, which is dependent on the deazaflavin cofactor F420 (25). In turn, F420, whose redox cycling requires the glucose-6-phosphate dehydrogenase Fgd1, is synthesized by FbiA, FbiB, and FbiC (5, 6, 38). The loss of any of these five enzymes leads to PA-824 resistance.
In contrast to Mycobacterium leprae, which lacks Ddn and is intrinsically resistant to PA-824 (26), the aforementioned enzymes are present in all members of the MTBC. However, they are not fully conserved, which might result in differential efficacy of PA-824 in different parts of the world (5, 7, 9, 16, 18).
To investigate this possibility in a systematic manner, we sequenced ddn and fgd1 in a reference collection of 65 strains that encompass all major phylogenetic lineages of the MTBC. Furthermore, we analyzed the correlation between particular single-nucleotide polymorphisms (SNPs) and the MICs in these strains. Moreover, we performed in silico modeling of the mutations in Fgd1 using the previously solved crystal structure of this protein (4). We included three PA-824-resistant positive controls (H37Rv-T3, H37Rv-5A1, and H37Rv-14A1) which had been previously analyzed genotypically and phenotypically (19, 25).
MATERIALS AND METHODS
Mycobacterial strains and growth conditions.
A total of 65 Mycobacterium strains (62 clinical isolates and 3 ATCC reference strains), including M. tuberculosis, M. africanum, M. bovis, M. caprae, M. pinnipedii, M. microti, and “M. canettii” strains, were used in this study (Table 1). All strains belonged to a reference collection comprising all major phylogenetic lineages of the MTBC and had been described earlier (41). Most of these were pansusceptible to standard antituberculosis drugs. Furthermore, three well-characterized PA-824-resistant control strains (H37Rv-T3, H37Rv-5A1, and H37Rv-14A1) were included (19, 25). Strains used for DNA isolation and MIC determination were cultivated on Löwenstein-Jensen agar slants.
Table 1.
Sequence results of fgd1 and ddn and drug susceptibility data of selected strains using the modified proportion method in BACTEC MGIT 960a
| Key | Species | Genotypeb | ddn (Rv3547) | fgd1 (Rv0407) | MIC (μg/ml) |
|---|---|---|---|---|---|
| 9679/00 | M. tuberculosis | H37Rv ATCC | 0.25 | ||
| 9564/00 | M. bovis | ATCC | ttT/ttC Phe320Phe | <0.0312 | |
| 9550/00 | M. africanum | ATCC (WA 2) | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | <0.0312 | |
| 9532/03 | M. tuberculosis | Haarlem | aAg/aTg Lys270Met | 0.25 | |
| 2336/02 | M. tuberculosis | Haarlem | aAg/aTg Lys270Met | 0.125 | |
| 4130/02 | M. tuberculosis | Haarlem | aAg/aTg Lys270Met | 0.125 | |
| 4850/03 | M. tuberculosis | EAI | ttT/ttC Phe320Phe | ||
| 1797/03 | M. tuberculosis | EAI | ttT/ttC Phe320Phe | ||
| 947/01 | M. tuberculosis | EAI | ttT/ttC Phe320Phe | ||
| 2333/99 | M. tuberculosis | Uganda I | |||
| 2169/99 | M. tuberculosis | Uganda I | |||
| 2201/99 | M. tuberculosis | Uganda I | |||
| 2191/99 | M. tuberculosis | Uganda II | |||
| 2253/99 | M. tuberculosis | Uganda II | |||
| 2176/99 | M. tuberculosis | Uganda II | |||
| 10493/01 | M. tuberculosis | Ghana | |||
| 2570/02 | M. tuberculosis | Ghana | |||
| 10469/01 | M. tuberculosis | Ghana | |||
| 5390/02 | M. tuberculosis | Cameroon | |||
| 5400/02 | M. tuberculosis | Cameroon | |||
| 1428/02 | M. tuberculosis | Cameroon | |||
| 1417/02 | M. tuberculosis | Cameroon | |||
| 7968/03 | M. tuberculosis | LAM | Cgg/Tgg Arg23Trp | 0.25 | |
| 8885/03 | M. tuberculosis | LAM | <0.0312 | ||
| 946/03 | M. tuberculosis | LAM | 0.0625 | ||
| 2151/03 | M. tuberculosis | S type | |||
| 2318/06 | M. tuberculosis | S type | |||
| 6411/05 | M. tuberculosis | S type | |||
| 4412/04 | M. tuberculosis | X type | |||
| 9953/04 | M. tuberculosis | X type | |||
| 8431/05 | M. tuberculosis | X type | |||
| 7746/01 | M. tuberculosis | Delhi/CAS | ttT/ttC Phe320Phe | 0.0625 | |
| 7936/01 | M. tuberculosis | Delhi/CAS | ttT/ttC Phe320Phe | 0.0625 | |
| 2637/02 | M. tuberculosis | Delhi/CAS | Gcg/Acg Ala111Thr | tcC/tcT Ser234Ser ttT/ttC Phe320Phe | 0.125 |
| 4445/02 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | ||
| 3256/02 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | ||
| 3329/02 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | ||
| 1934/03 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | 0.125 | |
| 12594/02 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | 0.125 | |
| 1500/03 | M. tuberculosis | Beijing | ttT/ttC Phe320Phe | 0.0625 | |
| 5434/02 | M. africanum | WA 1a | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | gcC/gcT Ala271Ala ttT/ttC Phe320Phe | 0.125 |
| 1449/02 | M. africanum | WA 1a | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | gcC/gcT Ala271Ala ttT/ttC Phe320Phe | |
| 1473/02 | M. africanum | WA 1a | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | gcC/gcT Ala271Ala ttT/ttC Phe320Phe | |
| 5432/02 | M. africanum | WA 1b | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | gGt/gCt Gly145Ala ttT/ttC Phe320Phe | 0.0625 |
| 10473/01 | M. africanum | WA 1b | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | ttT/ttC Phe320Phe | <0.0312 |
| 10494/01 | M. africanum | WA 1b | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | ttT/ttC Phe320Phe | 0.0625 |
| 1443/02 | M. africanum | WA 1b | ctG/ctT Leu48Leu Gac/Aac Asp113Asn | ttT/ttC Phe320Phe | |
| 10462/01 | M. africanum | WA 2 | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | <0.0312 | |
| 10476/01 | M. africanum | WA 2 | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | <0.0312 | |
| 10514/01 | M. africanum | WA 2 | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | ||
| 5468/02 | M. africanum | WA 2 | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | ||
| 10517/01 | M. africanum | WA 2 | Aag/Gag Lys296Glu ttT/ttC Phe320Phe | ||
| 4258/00 | M. bovis | Bovis | ttT/ttC Phe320Phe | ||
| 751/01 | M. bovis | Bovis | ttT/ttC Phe320Phe | ||
| 7540/01 | M. bovis | Bovis | ttT/ttC Phe320Phe | ||
| 9577/99 | M. caprae | Caprae | ttT/ttC Phe320Phe | ||
| 1694/00 | M. caprae | Caprae | ttT/ttC Phe320Phe | ||
| 8986/99 | M. caprae | Caprae | ttT/ttC Phe320Phe | ||
| 7011/02 | M. pinnipedii | Seal | Ctc/Gtc Leu90Val | ttT/ttC Phe320Phe | 0.0625 |
| 7739/01 | M. pinnipedii | Seal | Ctc/Gtc Leu90Val | ttT/ttC Phe320Phe | <0.0312 |
| 416/01 | M. microti | Llama | Ctc/Gtc Leu90Val | ttT/ttC Phe320Phe | |
| 1479/00 | M. microti | Vole | Ctc/Gtc Leu90Val | ttT/ttC Phe320Phe | |
| 3040/99 | M. canettii | −39 G/C gaC/gaT Asp69Asp gtT/gtC Val148Val | atG/atA Met208Ile ttT/ttC Phe320Phe | 8 | |
| 3041/99 | M. canettii | Cgg/Agg Arg23Arg gaC/gaT Asp69Asp gtT/gtC Val148Val | ttT/ttC Phe320Phe | 8 | |
| 3151/08 | M. canettii | Cgg/Agg Arg23Arg gaC/gaT Asp69Asp gtT/gtC Val148Val | ttT/ttC Phe320Phe | ||
| M. tuberculosis | H37Rv-T3c | Cag/Gag Gln88Glu | >32 | ||
| M. tuberculosis | H37Rv-5A1d | >32 | |||
| M. tuberculosis | H37Rv-14A1e | T deletion at nucleotide position 38 | >32 |
The strains analyzed comprise three PA-824-resistant strains (H37Rv-T3, H37Rv-5A1, and H37Rv-14A1) and 65 strains from a reference collection that encompasses all major phylogenetic lineages. Unless otherwise stated, the wild-type sequence was found. SNPs are shown in uppercase letters in the sequence data.
EAI, East African Indian; LAM, Latin American Mediterranean; WA, West African.
DNA isolation, PCR, and sequencing.
Genomic DNA was isolated as described previously (40). DNA amplification and subsequent sequencing were performed using the following primers: 5′-CGA GCG CAC CGA CCA GAG C-3′ (Rv3547_F) and 5′-GCA TGG CCC GCA GGT GGA CAA-3′ (Rv3547_R) for ddn and 5′-CGT GGC CGC GAG CGA GGT GAA-3′ (Rv0407_F) and 5′-CGC CCG AAC CGT CAA CAA CAC TGG-3′ (Rv0407_R) for fgd1. An additional sequencing primer, 5′-CGG AGT TCA AGG AGC GGT TCG-3′ (Rv0407_S1), was used for fgd1. PCRs were done using HotStarTaq DNA polymerase (Qiagen) under the following conditions: 95°C for 15 min, 94°C for 30 s, 64°C (ddn)/62°C (fgd1) for 30 s, 72°C for 1 min (ddn)/2 min (fgd1), and 72°C for 10 min. The second through fourth steps were repeated 30 times.
The PCR products thus obtained were sequenced using an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA) and an ABI Prism BigDye Terminator kit, version 3.1, according to the manufacturer's instructions. The sequence data were analyzed using the DNAStar Lasergene software package (version 8.0) with M. tuberculosis H37Rv (GenBank sequence accession number AL123456.2) as the reference.
Drug susceptibility testing.
PA-824 drug susceptibility testing was performed in the supranational reference laboratory in Borstel, Germany, using the modified proportion method in the Bactec MGIT 960 system (33). The PA-824 concentrations used were 1, 0.5, 0.25, 0.125, 0.0625, and 0.0312 μg/ml. The M. canettii strains and the PA-824-resistant positive controls were additionally tested at concentrations of 32, 16, 8, 4, and 2 μg/ml.
In silico modeling.
Our modeling pipeline consisted of three main steps and relied on the use of the MODELLER program (34). First, a multiple sequence alignment (see Fig. S1 in the supplemental material) of sequences homologous to the M. tuberculosis Fgd1 protein was prepared by querying the UniProt nonredundant sequence database using the wild-type sequence of Fgd1 with the profile_build command in the MODELLER-9v6 program (27, 42). Of the identified hits, only those that aligned to the query sequence with a sequence identity higher than 30% (P value of 0.0) and belonged to the Mycobacterium genus were selected. This yielded 21 homologous sequences, which were then realigned to the wild-type structure of M. tuberculosis Fgd1 using the expresso command of the T-Coffee program (2). Second, the MODELLER-9v6 program was used to build a 3-dimensional model for each of the five mutations identified in the Fgd1 protein (34). The crystallographic structure of M. tuberculosis Fgd1 was used as the structural template (4). All MODELLER parameters were kept at their default values. Third, for the native structure of Fgd1 and each of the five mutant models, a number of sequence and structure features were calculated as previously described (29).
RESULTS
Sequence analyses.
Our analysis of fgd1 and ddn in a total of 65 MTBC strains revealed 17 SNPs and one deletion that are listed in detail in Table 1. All strains except those belonging to the Euro-American superlineage (M. tuberculosis East African Indian, Delhi/CAS, and Beijing, M. bovis, M. africanum, M. caprae, M. pinnipedii, M. microti, and M. canettii) were characterized by a synonymous SNP in fgd1 (Phe320Phe). Further lineage-specific mutations in fgd1 or ddn were detected for M. tuberculosis Haarlem, both M. africanum lineages (West African [WA] 1 and 2), and the closely related M. pinnipedii and M. microti. In addition, one SNP in fgd1 (Ala271Ala) separated WA 1a and 1b strains, thereby supporting a further subclassification of this phylogenetic lineage. M. canettii was characterized by two synonymous mutations in fgd1 that were shared by all three strains (Asp69Asp and Val148Val) and an additional synonymous mutation that was present in only two strains (Arg23Arg). In addition, some strains (7968/03, 2637/02, 5432/02, and 3040/99) contained unique SNPs.
Of the three PA-824-resistant positive-control strains, H37Rv-5A1 did not contain a mutation in either gene, whereas H37Rv-T3 harbored a mutation in fgd1 (Gln88Glu) and H37Rv-14A1 had a 1-base-pair deletion in ddn.
MIC determination.
The MICs of the three PA-824-resistant positive controls were >32 μg/ml. The MICs of representatives of the wider MTBC diversity displayed considerable variability in their MICs, albeit at concentrations that were lower than or equal to the MIC of M. tuberculosis H37Rv (0.25 μg/ml). In marked contrast, M. canettii strains had a considerably higher MIC of 8 μg/ml (Table 1).
In silico modeling.
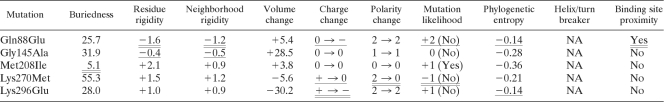
Predictions of the structural consequences of the point mutations were carried out for Fgd1 only, since no homologous structure was available for Ddn. Using the wild-type crystal structure of Fgd1 as a basis for comparison (4), four of the five mutations detected in clinical strains had only minor structural consequences (Table 2). Gly145Ala was a favorable mutation that did not occur in other mycobacteria, and the model predicted that the mutation had no effect on the function of the protein. Similarly, Met208Ile was a favorable mutation that was found in other mycobacteria. This mutation was predicted not to affect protein function even though it was located in the buried region of the protein. Despite being an unfavorable mutation that changed the polarity of the amino acid, Lys270Met was located in an exposed region of Fgd1 far from the F420 binding site. Lys296Glu also led to a change in the charge of the amino acid that was not observed in other mycobacteria. However, it was in an exposed region of the protein far from the binding site. Finally, of the five mutations, only Gln88Glu likely affected the F420 binding site, due to its close proximity in space. Moreover, this mutation did not occur in other mycobacteria and changed the charge in a well-defined and rigid region.
Table 2.
Structural properties of SNP models of the five Fgd1 nonsynonymous mutations detected in the 65 clinical strains and the PA-824-resistant control H37Rv-T3a
Unfavorable structural properties of the mutations in question are doubly underlined, intermediate properties are underlined, and the remaining properties were favorable. NA, not applicable.
DISCUSSION
Neither ddn nor fgd1 is essential for in vitro growth or survival in macrophages (32, 35, 36). Therefore, loss-of-function mutations anywhere in the gene can emerge under selection pressure, as exemplified by the ddn frameshift mutation in H37Rv-14A1. Similarly, SNPs along the whole length of the genes could potentially lead to PA-824 resistance. Indeed, we identified the hitherto unknown Gln88Glu mutation in fgd1 as the basis of resistance in the other PA-824-resistant mutant, H37Rv-T3 (19, 25). Our in silico modeling confirmed the significant impact of this mutation on the F420 binding site, thereby providing a plausible structural basis for drug resistance.
The sequence results of our reference collection were fully consistent with previous studies of the genetic diversity of the MTBC (5, 7, 16, 18). Indeed, we detected all known lineage-defining mutations in fgd1 and ddn and identified a novel marker for a subgroup among M. africanum WA 1 strains (1a, Ala271Ala). In addition, we identified some SNPs that occurred only in single strains, whereas other known SNPs (e.g., Arg64Ser in fgd1 [18]) were not found in our analysis, which highlights the need to test more strains in the future. Furthermore, this study was partially limited by the fact that we sequenced only two of the five genes known to be associated with resistance to PA-824. Given that the three remaining genes are not fully conserved either, the impact of their variability on intrinsic PA-824 resistance remains to be explored (5, 7, 9, 16, 18).
The maximal, averaged PA-824 plasma levels (Cmax) observed in healthy subjects during a study of a single dose of 1,500 mg and a multidose of 600 mg were 3 and 3.8 μg/ml, respectively (13). In smear-positive tuberculosis patients, the Cmax values at steady state were 2.4, 4.7, 7.1, and 7.5 μg/ml at once-daily doses of 200, 600, 1,000, and 1,200 mg, respectively (8; Global Alliance for TB Drug Development, personal communication).
In accordance with the results of previous studies, the three PA-824-resistant controls displayed MICs that were significantly higher than the plasma levels given above (19, 25). Apart from these, most MTBC strains tested were susceptible to PA-824 at MICs that were at least 9-fold lower than the concentrations achievable in patients. This is in line with prior reports (1, 8, 19, 21, 23, 26, 28, 38, 39). As a result, the nonsynonymous mutations detected in these strains should be regarded as neutral polymorphisms in future studies of PA-824 drug resistance.
However, we are the first to show that the MICs for M. canettii (8 μg/ml) are actually higher than all of the aforementioned Cmax values. In particular, they are more than 3-fold higher than the concentration reached by the 200 mg once-daily PA-824 dose which is currently being investigated in combination with moxifloxacin and pyrazinamide (14). This finding indicated that M. canettii strains might be intrinsically resistant to PA-824 at the doses evaluated so far. This would be in addition to the known intrinsic resistance of M. canettii against pyrazinamide (10, 22). Consequently, the combination of PA-824-moxifloxacin-pyrazinamide might result in monotherapy with only moxifloxacin, rendering this regimen unable to effectively treat TB caused by strains of this taxon (12).
These findings demand to be followed up on a number of fronts. First, the possibility exists that the strains tested are not representative of M. canettii and that the high MICs are limited to these two strains, given that this taxon is more diverse than other members of the MTBC (10, 17, 22). Additional previously isolated strains from other collections should therefore be tested to address this point. This is especially important because the genetic basis for the increased MICs remains unknown, as does the cause for the intrinsic resistance to pyrazinamide (10, 22). Indeed, our three M. canettii strains only had synonymous mutations in common, and even the lone upstream mutation and the nonsynonymous mutation (−39 G/C and Met208Ile) did not alter the MICs of the strain in question.
Second, it remains to be seen whether these increased MICs actually translate into intrinsic resistance in patients given that PA-824 concentrations in tissue are much higher than the concentrations in plasma, as observed in prior studies with rats (Global Alliance for TB Drug Development, personal communication). In theory, this question might have already been addressed during the past PA-824 clinical trials. Should the strains from these trials for which the MICs are already known still be available, their respective phylogenetic lineages should be determined (8). In practice, however, the past trials in South Africa probably did not feature M. canettii strains, given that this taxon is almost exclusively limited to the Horn of Africa, although more accurate data concerning its prevalence and spread are required (10, 17, 18, 22, 41). As a result, even active screening for patients with TB caused by M. canettii with the aim of enrolling them in trials will probably not identify sufficient numbers of patients unless the trials are conducted in the Horn of Africa. Therefore, animal models might remain as the only realistic avenue to study M. canettii in vivo.
The results of our study are also of relevance for OPC-67683, the second nitroimidazole in phase II trials (3, 12). The only genetically characterized OPC-67683-resistant mutants that have been tested to date contain mutations in ddn (see the comments in the supplemental material), but cross-resistance probably extends to the remaining four PA-824 resistance loci (3, 12, 19, 28). If complete cross-resistance is assumed, then our conclusions can be cautiously extended to OPC-67683 (i.e., most MTBC strains should be susceptible to OPC-67683). But given that OPC-67683 is more potent than PA-824 (19, 28), its plasma levels might be high enough to overcome the higher MICs of M. canettii. Despite having sampled the global diversity of the MTBC in a variety of countries, the clinical trials for this antibiotic probably did not include M. canettii either (12, 18). As a result, the MICs of OPC-67683 should be determined in vitro.
Similarly, screening of reference collections comparable to ours is required for metronidazole, the third nitroimidazole in phase II clinical trials (12). The same applies to further nitroimidazoles that are in preclinical development. The latter lead compounds display better in vitro activities than PA-824 and might therefore be able to overcome the potential intrinsic resistance (15).
In conclusion, the genetic diversity within the MTBC plays a role in the use of current antibiotics, such as pyrazinamide, as well as new drug candidates in clinical trials (10, 22). This variation has to be catalogued in a systematic manner and correlated with MICs. This should become a required part of preclinical trials, since the clinical trials themselves cannot be expected to include representatives of all MTBC lineages. Moreover, the identity of all strains from patients should be routinely determined as an additional control in future clinical trials (30). Importantly, the fact that M. canettii was found to be potentially intrinsically resistant to PA-824, despite the lack of a clear genetic basis for this phenotype, exemplifies the possibility that other MTBC lineages might be intrinsically resistant to other drug candidates.
Supplementary Material
ACKNOWLEDGMENTS
H. Boshoff and C. Barry III provided the PA-824 for this study and commented on the manuscript, for which we are very grateful. We thank A. Diacon, N. Erondu, and A. Ginsberg (Global Alliance for TB Drug Development) for sharing and discussing the PA-824 clinical trial data and M. Matsumoto for sharing the precise identity of the OPC-67683 resistance mutations. We also thank T. Ubben, L. Dost, and I. Razio (Borstel, Germany) for excellent technical assistance and J. Parkhill for helpful discussions of the manuscript. G. Barbour and S. Jones also contributed to this work.
S. Niemann was supported by the European Union TB-PAN-NET (FP7-223681) project and M. A. Marti-Renom by the Spanish Ministerio de Ciencia e Innovación (BFU2010-19310) and Era-Net Pathogenomics (PIM2010EPA-00719). C. Köser is a recipient of a Gates Cambridge Scholarship and received additional funding from the Cambridge Philosophical Society, the Cambridge European Trust, and Clare Hall, Cambridge.
The authors declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 19 September 2011.
REFERENCES
- 1. Ahmad Z., et al. 2011. PA-824 exhibits time-dependent activity in a murine model of tuberculosis. Antimicrob. Agents Chemother. 55:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Armougom F., et al. 2006. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34:W604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barry C. E., III, Blanchard J. S. 2010. The chemical biology of new drugs in the development for tuberculosis. Curr. Opin. Chem. Biol. 14:456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bashiri G., Squire C. J., Moreland N. J., Baker E. N. 2008. Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J. Biol. Chem. 283:17531–17541 [DOI] [PubMed] [Google Scholar]
- 5. Choi K., Kendrick N., Daniels L. 2002. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F(420) and FO biosynthesis. J. Bacteriol. 184:2420–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi K. P., Bair T. B., Bae Y. M., Daniels L. 2001. Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG. J. Bacteriol. 183:7058–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Comas I., Homolka S., Niemann S., Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diacon A. H., et al. 2010. Early bactericidal activity and pharmacokinetics of PA-824 in smear-positive tuberculosis patients. Antimicrob. Agents Chemother. 54:3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Domenech P., Kolly G. S., Leon-Solis L., Fallow A., Reed M. B. 2010. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J. Bacteriol. 192:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabre M., et al. 2010. Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect. Genet. Evol. 10:1165–1173 [DOI] [PubMed] [Google Scholar]
- 11. Farmer P. E., Nizeye B., Stulac S., Keshavjee S. 2006. Structural violence and clinical medicine. PLoS Med. 3:e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ginsberg A. M. 2010. Drugs in development for tuberculosis. Drugs 70:2201–2214 [DOI] [PubMed] [Google Scholar]
- 13. Ginsberg A. M., Laurenzi M. W., Rouse D. J., Whitney K. D., Spigelman M. K. 2009. Safety, tolerability, and pharmacokinetics of PA-824 in healthy subjects. Antimicrob. Agents Chemother. 53:3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Global Alliance for TB Drug Development 2011. Evaluation of early bactericidal activity in pulmonary tuberculosis with(J-M-Pa-Z), clinical trial. ClinicalTrials.gov identifier NCT01215851. Global Alliance for TB Drug Development, New York, NY: http://www.clinicaltrials.gov/ct2/show/NCT01215851 Accessed 23 January 2011 [Google Scholar]
- 15. Global Alliance for TB Drug Development 2011. Nitroimidazoles. Global Alliance for TB Drug Development, New York, NY: http://www.tballiance.org/new/portfolio/html-portfolio-item.php?id=1 Accessed 14 January 2011 [Google Scholar]
- 16. Gutacker M. M., et al. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutierrez M. C., et al. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 1:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hershberg R., et al. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6:e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hurdle J. G., et al. 2008. A microbiological assessment of novel nitrofuranylamides as anti-tuberculosis agents. J. Antimicrob. Chemother. 62:1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hutson S. 2010. Half-century-old TB drugs get a facelift in new cocktails. Nat. Med. 16:1346. [DOI] [PubMed] [Google Scholar]
- 21. Ji B., et al. 2006. In vitro and in vivo activities of rifampin, streptomycin, amikacin, moxifloxacin, R207910, linezolid, and PA-824 against Mycobacterium ulcerans. Antimicrob. Agents Chemother. 50:1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koeck J. L., et al. 2011. Clinical characteristics of the smooth tubercle bacilli “Mycobacterium canettii” infection suggest the existence of an environmental reservoir. Clin. Microbiol. Infect. 17:1013–1019 [DOI] [PubMed] [Google Scholar]
- 23. Lenaerts A. J., et al. 2005. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob. Agents Chemother. 49:2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manjunatha U., Boshoff H. I., Barry C. E., III 2009. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun. Integr. Biol. 2:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manjunatha U. H., et al. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manjunatha U. H., et al. 2006. Mycobacterium leprae is naturally resistant to PA-824. Antimicrob. Agents Chemother. 50:3350–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marti-Renom M. A., Madhusudhan M. S., Sali A. 2004. Alignment of protein sequences by their profiles. Protein Sci. 13:1071–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsumoto M., et al. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3:e466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mirkovic N., Marti-Renom M. A., Weber B. L., Sali A., Monteiro A. N. 2004. Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Res. 64:3790–3797 [DOI] [PubMed] [Google Scholar]
- 30. Nahid P., et al. 2010. Influence of M. tuberculosis lineage variability within a clinical trial for pulmonary tuberculosis. PLoS One 5:e10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nathan C. 2008. Microbiology. An antibiotic mimics immunity. Science 322:1337–1338 [DOI] [PubMed] [Google Scholar]
- 32. Rengarajan J., Bloom B. R., Rubin E. J. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rüsch-Gerdes S., Pfyffer G. E., Casal M., Chadwick M., Siddiqi S. 2006. Multicenter laboratory validation of the BACTEC MGIT 960 technique for testing susceptibilities of Mycobacterium tuberculosis to classical second-line drugs and newer antimicrobials. J. Clin. Microbiol. 44:688–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sali A., Blundell T. L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 35. Sassetti C. M., Boyd D. H., Rubin E. J. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 36. Sassetti C. M., Rubin E. J. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh R., et al. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stover C. K., et al. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962–966 [DOI] [PubMed] [Google Scholar]
- 39. Tyagi S., et al. 2005. Bactericidal activity of the nitroimidazopyran PA-824 in a murine model of tuberculosis. Antimicrob. Agents Chemother. 49:2289–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Soolingen D., Hermans P. W., de Haas P. E., Soll D. R., van Embden J. D. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wirth T., et al. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu C. H., et al. 2006. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34:D187–D191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.