Abstract
The extracellular matrix protein adhesin A (EmaA) of the Gram-negative bacterium Aggregatibacter actinomycetemcomitans is a fibrillar collagen adhesin belonging to the family of trimeric autotransporters. The protein forms antenna-like structures on the bacterial surface required for collagen adhesion. The 202-kDa protein monomers are proposed to be targeted and translocated across the inner membrane by a long signal peptide composed of 56 amino acids. The predicted signal peptide was functionally active in Escherichia coli and A. actinomycetemcomitans using truncated PhoA and Aae chimeric proteins, respectively. Mutations in the signal peptide were generated and characterized for PhoA activity in E. coli. A. actinomycetemcomitans strains expressing EmaA with the identical mutant signal peptides were assessed for cellular localization, surface expression, and collagen binding activity. All of the mutants impaired some aspect of EmaA structure or function. A signal peptide mutant that promoted alkaline phosphatase secretion did not allow any cell surface presentation of EmaA. A second mutant allowed for cell surface exposure but abolished protein function. A third mutant allowed for the normal localization and function of EmaA at 37°C but impaired localization at elevated temperatures. Likewise, replacement of the long EmaA signal peptide with a typical signal peptide also impaired localization above 37°C. The data suggest that the residues of the EmaA signal peptide are required for protein folding or assembly of this collagen adhesin.
INTRODUCTION
An important step in tissue colonization for many bacteria is binding to the extracellular matrix (54). Surface proteins are essential mediators for establishing these infectious foci. The extracellular matrix protein adhesin A (EmaA) is critical for the binding of the oral and systemic pathogen Aggregatibacter actinomycetemcomitans to fibrillar collagens (28). A. actinomycetemcomitans is a Gram-negative, capnophilic bacterium associated with both adult and juvenile forms of periodontal disease (8, 44, 59). This bacterium is also associated with nonoral diseases including but not limited to pneumonia, bone disease, soft tissue abscesses, and infective endocarditis (6, 27, 32, 40). It has been shown that the loss of EmaA activity impairs the colonization of traumatized heart valves in a rabbit infective endocarditis model by A. actinomycetemcomitans (48).
EmaA belongs to a specific family of autotransporter proteins and is secreted via the type V secretion pathway (17, 28). In general, autotransporters are targeted to the membrane by a signal peptide and translocated across the inner membrane using the Sec translocon (12, 17). Once in the periplasmic space, the carboxyl terminus of the protein is proposed to form an outer membrane pore for secretion of the passenger domain through a yet undefined mechanism (10, 16). Following translocation across the outer membrane, the protein is either retained on the surface or proteolytically released into the milieu (12). EmaA and members of the type Vc secretion pathway proteins differ from the traditional autotransporters by requiring three monomers to form the outer membrane pore for secretion (17). This adhesin is synthesized as 202-kDa monomers, which trimerize to form surface antenna-like structures required for collagen binding (42). Inactivation of emaA results in a bacterial strain that does not display surface structures and reduces the ability of the bacterium to bind to both purified and tissue collagens (42, 48). The biological activity of the EmaA structure has been assigned to the most distal domain of the antenna-like structures, which is composed of the amino termini of the monomers, amino acids 70 to 386 (58). Recently, the three-dimensional architecture of the functional domain of this adhesin has been described and is composed of three subdomains (57, 58). In addition, oligosaccharides are suggested to be required for collagen binding, which are related to the O polysaccharide (O-PS) of the lipopolysaccharide (LPS) of A. actinomycetemcomitans (49).
Targeting to and translocation of the EmaA monomers across the inner membrane are predicted to be mediated by an amino-terminal signal peptide (28). The length of the predicted EmaA signal peptide (56 amino acids) is much greater than that of typical signal peptides, which are composed of up to 25 amino acids (52). These long signal peptides are found predominantly associated with proteins secreted through the autotransporter, type V protein secretion pathway; long signal peptides are present in all of the serine protease autotransporters of members of the Enterobacteriaceae family (SPATEs) (47), some proteins of the two-partner secretion (TPS) pathway (19), select trimeric autotransporters (46), as well as the eukaryotic proteins interleukin 15 (IL-15) and plasminogen activator inhibitor 2 (3, 26).
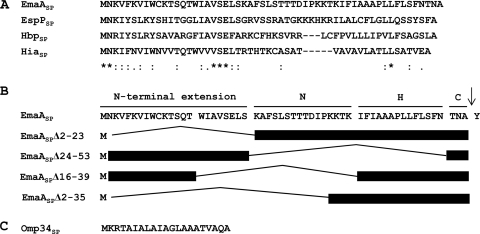
Amino-terminal signal peptides exhibit limited sequence similarity but are composed of clusters of charged or hydrophobic amino acids that are required for interaction with the protein secretory machinery in the cytoplasm (10, 51). Typical signal peptides are divided into three regions, with a variable number of amino acids. An uncommonly high number of positively charged amino acids present after the start methionine constitute the N region, followed by a region of hydrophobic amino acids (H region) adjacent to a sequence containing the cleavage site for the inner membrane-bound signal peptidase (C region). The C region usually contains small, slightly polar amino acids at the −1 and −3 positions of the signal peptidase cleavage site (53). The carboxyl-terminal half of long signal peptides contains the canonical N, H, and C regions of typical signal peptides but displays high sequence variability (47). Conversely, the amino-terminal amino acids contain a unique, highly conserved sequence motif, referred to as the N-terminal extension or extended signal peptide region (ESPR) (Fig. 1A and B) (11, 47). Some long signal peptides may be composed of two divergent NH regions followed by a C region, denoted N1H1N2H2C, with little sequence similarity between the individual N and H regions (11, 17). The role of the N-terminal extension remains to be elucidated.
Fig. 1.
Signal peptides of selected Gram-negative bacterial proteins. (A) Sequence alignment of long signal peptides of proteins from different organisms. Alignments were generated using ClustalW2 (7). Identical amino acids (*), highly conserved amino acids (:), and weakly conserved amino acids (.) are indicated below the sequence alignment. Gaps introduced to maximize sequence alignment are indicated by the dashes. (B) Predicted domain structure of the EmaA signal peptide and schematics of the in-frame EmaA signal peptide deletion constructs used in this study. The arrow indicates the predicted signal peptidase cleavage site between amino acid 56 (Ala) and 57 (Tyr). (C) Sequence of the typical signal peptide of Omp34 of A. actinomycetemcomitans.
In this study, we demonstrate that the first 56 amino acids of the EmaA sequence are required for protein secretion. The results of amino acid deletion analysis of the signal peptide suggest that the residues are required for proper protein folding and assembly of this collagen adhesin. Furthermore, we have demonstrated that part of the long EmaA signal peptide is important for the secretion of this virulence determinant in response to changes in growth conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless noted, all A. actinomycetemcomitans strains were grown statically in 3% Trypticase soy broth supplemented with 0.6% yeast extract (TSBYE) in a 10% CO2 humidified incubator at 37°C. Antibiotics were added to a final concentration of 50 μg/ml kanamycin or 1 μg/ml chloramphenicol for A. actinomycetemcomitans, when necessary. All Escherichia coli strains were grown with agitation in Luria-Bertani broth (LB) at 37°C containing 50 μg/ml kanamycin, 20 μg/ml chloramphenicol, 50 μg/ml ampicillin, or 500 μg/ml erythromycin, as appropriate.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description or relevant genotype or phenotype | Reference or source |
|---|---|---|
| A. actinomycetemcomitansstrains | ||
| VT1169 | Wild-type strain, a derivative of SUNY465 | 31 |
| KM73 | emaA mutant strain | 28 |
| VT1565 | aae mutant strain | 41 |
| KM333 | In-frame deletion of emaA gene region corresponding to deletion of amino acids 16 to 39 of EmaA protein | This study |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsd RMS-mcr BC) φ80dlacZΔM15 Δlac X74 end A1 rec A1 deo R Δ(ara leu)7697 ara D139 gal U gal K nup G rps L λ− | 15 |
| DH5α λ pir | endA1 hsdR17(r− m+) supE44 thi-1 recA1 gyrA1(Nalr) relA1Δ(lacIZYA-argF)U169 deo[φ80 ΔlacD(lacZ)M15] pir R6K | |
| SM10 λ pir | thi-1 thr leu tonA lacY supE recA::RP4-2Tc::Mu l pir R6K | 39 |
| CC118 | phoA20 thi-1 rspE rpoB argE(Am) recA1 | 18 |
| Plasmids | ||
| pKM1 | Shuttle vector; Kanr | 42 |
| pKM2 | Shuttle vector; Cmr | 14 |
| pVT1460 | Mobilization plasmid | 29 |
| pVT1566 | aae gene sequence cloned into pGEM-T Easy | 41 |
| pKM9 | Upstream promoter region and emaA in pKM1 | 42 |
| pQE30 | Intermediate cloning vector; Ampr | Qiagen |
| pHRM104 | Plasmid containing truncated phoA | 17 |
| pQEmaA | Upstream promoter region and emaA in pQE30 | This study |
| pKMΔ2-23 | In-frame deletion of bp 4 to 69 of emaA in pKM9 | This study |
| pKMΔ2-35 | In-frame deletion of bp 4 to 105 of emaA in pKM9 | This study |
| pKMΔ24-53 | In-frame deletion of bp 70 to 159 of emaA in pKM9 | This study |
| pKMΔ16-39 | In-frame deletion of bp 46 to 117 of emaA in pKM9 | This study |
| pKMOmp34SP | emaA signal peptide replaced by omp34 signal peptide in pKM9 | This study |
| pKMEmaASP-Aae | aae containing the emaA signal peptide and upstream promoter region in pKM2 | This study |
| pHRMEmaASP | emaA signal peptide and upstream promoter region in pHRM104 | This study |
| pHRMEmaASPΔ2-56 | In-frame deletion of bp 4 to 168 of pHRMEmaASP | This study |
| pHRMEmaASPΔ2-23 | In-frame deletion of bp 4 to 69 of pHRMEmaASP | This study |
| pHRMEmaASPΔ2-35 | In-frame deletion of bp 4 to 105 of pHRMEmaASP | This study |
| pHRMEmaASPΔ24-53 | In-frame deletion of bp 70 to 159 of pHRMEmaASP | This study |
| pHRMEmaASPΔ16-39 | In-frame deletion of bp 46 to 117 of pHRMEmaASP | This study |
| pHRMHiaSP | hia signal peptide and emaA upstream promoter region in pHRM104 | This study |
For some experiments, bacteria were adapted for growth at 39°C. Bacteria, from frozen stocks, were inoculated on TSBYE agar and grown at 39°C in 10% CO2. Colonies were subsequently inoculated into prewarmed TSBYE broth and grown statically at 39°C. For heat shock experiments, the cells were initially grown in TSBYE at 37°C overnight. The cells were diluted, grown at 37°C for 30 min in 10% CO2, sealed, and incubated at 42°C for 3 h before analysis. Bacterial growth at 39°C and 42°C was comparable to the growth of cells grown at 37°C for the stated periods of time.
In-frame deletion constructs.
A shuttle plasmid (pKM9) containing the full-length emaA sequence and 500 bp upstream of the start methionine (GenBank accession number AY344064) was used to generate in-frame deletion constructs utilizing unique 5′ SphI and 3′ KpnI (within the emaA sequence) restriction sites for directional cloning (58). The deletion constructs were generated by overlapping PCR methodology. Three rounds of PCR were completed. The first round utilized a forward primer (CBP1-5′up) and a reverse primer designed to contain the complementary sequence flanking the sequence of interest to encompass the 5′ SphI site. The second round utilized a forward primer complementary to the sequence of interest and the reverse primer CBP1-3131Rev to generate a product containing the unique 3′ KpnI restriction site (Table 2). The PCR products were purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) and used as the template for the final round of PCR using the primer set CBP1-5′up and CBP1-3131Rev. The final PCR product was purified, treated with the respective enzymes, and ligated with purified pQEmaA (Table 1). The ligation product was transformed into E. coli DH10B cells by electroporation (45), and ampicillin-resistant colonies were selected. Plasmid DNA was digested with SphI and SmaI (located at the 3′ end of the emaA gene), ligated with pKM1 (Table 1) previously digested with SphI and HincII and transformed into E. coli DH10B. Kanamycin-resistant colonies were selected, and the purified plasmid containing the modified emaA sequence was transformed into the emaA mutant strain of A. actinomycetemcomitans by electroporation as described previously (45). All PCR constructs were confirmed by DNA sequence analysis performed at the Vermont Cancer Center DNA Analysis Facility, University of Vermont.
Table 2.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′-3′)b |
|---|---|
| CBP1-5′up | ACATGCATGCAACAAATCGCCGTCATCGCC |
| CBP1-3131Rev | GACTGCTAAATTCTTTCCTGCC |
| EmaAΔ2-23For | CAAAAAGGAAAACATAAGATGAAAGCTTTTTCCCTTTCTACCAC |
| EmaAΔ2-23Rev | GTGGTAGAAAGGGAAAAAGCTTTCATCTTATGTTTTCCTTTTTG |
| EmaAΔ24-53For | GAACTATCTTTTAATACCAACGCTTACATTGCTATAGG |
| EmaAΔ24-53Rev | CCTATAGCAATGTAAGCGTTGGTATTAAAAGATAGTTC |
| EmaAΔ16-39For | GGTGTAAAACATCTCAGACAATATTCATTGCTGCAGCCCCG |
| EmaAΔ16-39Rev | CGGGGCTGCAGCAATGAATATTGTCTGAGATGTTTTACACC |
| omp34SPFor | GCACCACAAGCAAACACT |
| omp34SPRev | CACTACGAATTAAAGCGG |
| emaAP-omp34SPFor | CAAAAAGGAAAACATAAGATGAAAAGAACTGCAATC |
| emaAP-omp34SPRev | GATTGCAGTTCTTTTCATCTTATGTTTTCCTTTTTG |
| omp34SP-emaAFor | GCAACAGTAGCACAGGCATACATTGCTATAGGTTCT |
| omp34SP-emaARev | AGAACCTATAGCAATGTATGCCTGTGCTACTGTTGC |
| emaASP-aaeFor | TCCTTTAATACCAACGCTTCAGAGTTTAATGCTCAAATAAATAAT |
| emaASP-aaeRev | ATTATTTATTTGAGCATTAAACTCTGAAGCGTTGGTATTAAAGGA |
| aaeBamFor | GGATCCTTCAGAGTTTAATGCTCAA |
| aaeSacRev | GAGCTCTTACCAGTAGTAATTCAG |
| HiaSP-EmaA5′For | CAAAAAGGAAAACATAAGATGAACAAAATTTTTAACG |
| HiaSP-EmaA5′Rev | CGTTAAAAATTTTGTTCATCTTATGTTTTCCTTTTTG |
| HiaSP-EmaA3′For | GTCCGCAACGGTTGAGGCGTTACATTGCTATAGGTTCTG |
| HiaSP-EmaA3′Rev | CAGAACCTATAGCAATGTAACGCCTCAACCGTTGCGGAC |
| EmaASPBamHIRev | GGATCCAGCGTTGGTATTAAA |
| EmaAPromoterBamHIRev | GGATCCCATCTTATGTTTTCCTTTTTG |
| EmaAΔ24-53BamHIRev | GGATCCAGATAGTTCAGATACGGC |
| Omp34SPBamHIRev | GGATCCTGCCTGTGCTACTGTTGCTGC |
| HiaSPBamHIRev | GGATCCCGCCTCAACCGTTGCGG |
| PhoA396Rev | AATATCGCCCTGAGCAGCC |
| CoA1347StuIFor | GATGCAGGCCTCGACGGCAATTTATACATC |
| CBP1-7StuIRev | GGAGAAGGCCTTTGACGCATCATCGCAAG |
| CoA-aad95′For | GTATAACTAAATGATTCATCATCGATTTTCGTTCGTGAATAC |
| CoA-aad95′Rev | GTATTCACGAACGAAAATCGATGATGAATCATTTAGTTATAC |
| aad9-EmaAFor | CAATAAACCTTGCATATGGATTCATCATAAAAGTGC |
| aad9-EmaARev | GCACTTTTATGATGAATCCATATGCAAGGTTTATTG |
| Spc2Rev | CTCTTGCCAGTCACGTTACG |
| 16SrRNAfor | GAACCTTACCTACTCTTGACATCC |
| 16SrRNArev | GGACTTAACCCAACATTTCACAAC |
| 16SrRNAprobe | 6-FAM-CTGACGACAGCCATGCAGCACCTG-BHQ-1 |
| emaAstalkfor | GGTCAATAACGACGGTGTTACG |
| emaAstalkrev | TTCCCTTTCGCAACGTTAGC |
| emaAstalkprobe | 6-FAM-CGGTCCAAGCATGACAAGCCACGG-BHQ-1 |
Forward (For) and reverse (rev) oligonucleotide primers and probes.
6-FAM, 6-carboxyfluorescein; BHQ-1, black hole quencher 1.
Construction and analysis of phoA fusions.
Alkaline phosphatase fusion constructs were generated using pHRM104, a plasmid containing a truncated gene for alkaline phosphatase (phoA) that lacks a functional signal sequence, as the backbone (36). The emaA signal peptide in-frame deletion constructs containing the emaA promoter region were amplified with engineered BamHI sites (Table 2). The purified PCR products were treated with BamHI and ligated with pHRM104 previously digested with BamHI. The ligation mixture was transformed into electrocompetent E. coli CC118 cells with mutant phoA genes, and erythromycin-resistant transformants were selected. The orientation of the insert was verified by PCR using the requisite primer set (Table 2). The hia signal peptide-phoA fusion was kindly provided by J. St. Geme, Duke University Medical Center, Durham, North Carolina (46).
PhoA activity was determined based on the colorimetric assay of Brickman and Beckwith (5). Data were collected from three individual experiments performed in triplicate and statistically analyzed for significance using Student's t test for statistical significance (P < 0.05).
Heterologous signal peptide fusion constructs.
Two heterologous signal peptide fusions using A. actinomycetemcomitans outer membrane proteins were developed in this investigation. The nucleotide sequences corresponding to the signal peptides of the outer membrane protein 34 (Omp34) (GenBank accession number AB015936) (55) and EmaA was generated by overlapping PCR methodology. The omp34 signal peptide sequence was generated by PCR using the requisite primer set (Table 2) and genomic DNA as the template. The 500-bp sequence upstream of the emaA start codon and the 5′ end of the emaA gene, after the signal peptide sequence, were amplified (Table 2) and purified. A mixture of the three products was used as template with the requisite primer set (Table 2), and the PCR product was treated with SphI and KpnI, ligated with pQEmaA previously digested with the corresponding enzymes, and transformed into E. coli DH10B cells.
The signal peptide of EmaA was fused to the epithelial cell adhesin, Aae (GenBank accession number AY262734) (41), with the endogenous signal peptide deleted, by overlapping PCR methodology. The emaA upstream sequence and the sequence for the signal peptide were amplified using the requisite primers, one of which contains the complementary sequence for the aae gene downstream of the signal peptide sequence. In a separate reaction, the aae sequence from bp 82 of the coding sequence (minus the signal peptide sequence) was amplified (Table 2). The purified PCR products were used as the template with the requisite primers (Table 2) to generate the fusion product. The purified product was treated with SphI and SacI and ligated with the empty plasmid (pKM2) previously treated with the same enzymes. The ligation product was transformed into the aae mutant strain VT1565 (41).
Surface detection of Aae.
An enzyme-linked immunosorbent assay (ELISA) was developed based on a modified version of the method of Mintz and Fives-Taylor (30). Briefly, 1 × 108 CFU of mid-log-phase bacterial cells were adsorbed to microtiter plate wells, and surface-bound Aae was localized by the addition of anti-Aae polyclonal antiserum. Immunoreactive complexes were detected by the addition of enzyme-conjugated goat anti-rabbit serum (Sigma-Aldrich, Inc., St. Louis, MO) in the presence of a colorimetric substrate. Data were collected from three individual experiments in triplicate and statistically analyzed for significance using Student's t test for significance (P < 0.05).
Generation of a chromosomal in-frame EmaA signal peptide deletion mutant.
A mutant with a chromosomal deletion of nucleotides 46 to 117 of the emaA coding sequence, corresponding to amino acids 16 to 39, was generated allelic replacement of the wild-type sequence with a DNA fragment containing a selectable marker 500 bp upstream of the deleted sequence. The DNA fragment for homologous recombination was generated using overlapping PCR methodology. A 500-bp DNA fragment of the coenzyme A (CoA) ligase gene, which is found upstream of the emaA sequence, was amplified with an engineered 5′ sequence of the aad9 selectable marker (GenBank accession number M69221) (Table 2). The aad9 selectable marker was amplified to generate DNA with ends complementary to the CoA ligase and the intergenic sequence of emaA (Table 2). A plasmid containing the in-frame deletion corresponding to amino acids 16 to 39 was used as the template to generate the third DNA fragment for overlapping PCR (Table 2). The PCR products were purified, combined, and used as the template for PCR using the requisite primers (Table 2). The overlapping PCR product was cloned into the TA cloning vector TOPO (Invitrogen, Carlsbad, CA). The 2.7-kb fragment was excised from the plasmid by digestion with EcoRI, purified, and ligated with the pVT1460, previously treated with EcoRI, for conjugation as described previously (29).
Quantitative real-time PCR.
Total bacterial RNA was isolated using the Qiagen RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol and used for reverse transcription using SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Quantitative real-time PCR was performed as described previously (57) in the DNA Analysis Facility, Vermont Cancer Center, at the University of Vermont. The primers and the TaqMan fluorogenic probes (Sigma Chemical Co.) used are present in Table 2. The expression of emaA was normalized to expression of the endogenous 16S rRNA for variation in RNA quantity and quality.
The relative quantification of target gene expression was performed using the comparative cycle threshold (CT) method (1a). The emaA mutant strain transformed with pKM9 was chosen as the calibrator. Results are presented as the means ± standard deviations of three independent experiments.
Transmission electron microscopy.
Bacterial samples were prepared by the method of Ruiz et al. (42) using Nano-W (Nanoprobes, Yaphank, NY) as the staining agent. Data collection was carried out using a Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 cathode, a 14-μm, 2,048- by 2,048-pixel charge-coupled-device (CCD) camera (TVIPS, Gauting, Germany) and a dual-axis tilt tomography holder (Fischione, Export, PA), operating at 100 kV. Micrographs were recorded using the CCD camera at a nominal magnification of ×52,000, which corresponds to a 0.25-nm pixel size on the specimen scale.
Isolation of bacterial membranes and aggregated proteins.
Bacterial membranes were prepared following the protocol described by Mintz (28). The protein concentration was determined spectrophotometrically at a wavelength of 280 nm following the addition of sodium dodecyl sulfate (SDS) to a final concentration of 2% (wt/vol).
Aggregated proteins in the membrane fraction samples were isolated according to the protocol of Tomoyasu et al. (50). Briefly, membrane fraction samples were treated with 10% (vol/vol) NP-40, and the aggregated proteins were isolated by centrifugation and analyzed for EmaA aggregates by immunoblotting.
Immunoblot analysis.
The amount of EmaA synthesized by the bacterial strains was analyzed by immunodot blot analysis of membrane fragments following the procedure of Yu et al. (57) using a monoclonal antibody specific for the stalk region of EmaA. The nitrocellulose membranes were exposed to photographic film or imaged using a Bio-Rad Molecular Imager Gel Doc XR+ system (Bio-Rad Laboratories). The intensity of the dots was quantified using the Bio-Rad Quantity One software.
Membrane localization of Aae was determined by immunoblotting of SDS-polyacrylamide gels transferred to nitrocellulose membranes using polyclonal antisera raised against a recombinant protein corresponding to the passenger domain of Aae (41).
The presence of PhoA in the bacterial whole-cell lysates was determined by immunoblotting of SDS-polyacrylamide gels transferred to nitrocellulose membranes using rabbit anti-PhoA antiserum (Rockland Immunochemicals Inc., Gilbertsville, PA). The immune complexes were detected by horseradish peroxidase-conjugated goat anti-rabbit antibodies and exposed to photographic film.
Collagen binding assay.
Collagen binding activity was determined as described previously (58). Data were collected from three individual experiments in triplicate and statistically analyzed for significance using Student's t test (P < 0.05) or two-way analysis of variance (ANOVA) where appropriate.
RESULTS
The first 56 amino acids of EmaA act as a signal peptide for inner membrane translocation in E. coli.
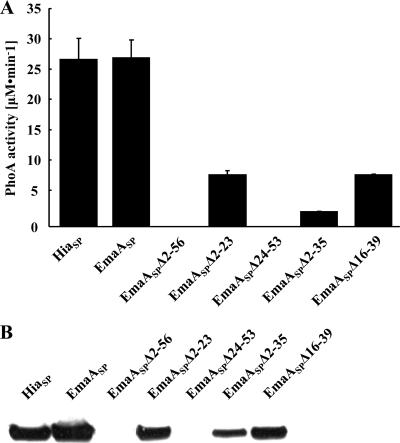
A signal peptidase cleavage site between amino acids 56 and 57 of the EmaA protein sequence (Fig. 1B) is predicted by the Signal P algorithm (4, 33, 34). The membrane targeting and translocation activity of this sequence was determined in E. coli using a phoA reporter construct, which lacks the native signal peptide and promoter (36, 46). PhoA activity is detected only when the enzyme is translocated across the bacterial inner membrane (5). Alkaline phosphatase activity was clearly associated with the emaASP-phoA construct as shown in Fig. 2. The activity of the strain expressing EmaASP (EmaA signal peptide corresponding to amino acids 1 to 56) was comparable to the activity associated with the strain expressing the 49-amino-acid signal peptide of the Haemophilus influenzae trimeric autotransporter epithelial cell adhesin Hia (46) fused to PhoA (HiaSP) used as a positive control. These activities are in contrast to the lack of enzymatic activity associated with the construct that contains the promoter but lacks the first 56 amino acids of the EmaA sequence (EmaASPΔ2-56). The presence of phosphatase activity in the EmaASP-expressing strain suggested that the first 56 amino acids of EmaA were sufficient to mediate the targeting and translocation of PhoA across the bacterial inner membrane.
Fig. 2.
Alkaline phosphatase activity of signal peptide constructs in E. coli. Plasmids were constructed with the emaA promoter adjacent to DNA sequences that encode the entire signal peptide or regions of the emaA signal peptide fused to a truncated form of phoA (pHRM104). All constructs were transformed into strain CC118, an E. coli phoA mutant strain. (A) PhoA enzymatic activity represented as micromoles of para-nitrophenyl generated per minute. HiaSP, signal peptide from the epithelial cell adhesin Hia of H. influenzae (amino acids 1 to 49); EmaASP, EmaA signal peptide corresponding to amino acids 1 to 56; EmaASPΔ2-56, EmaA signal peptide with amino acids 2 to 56 deleted; EmaASPΔ2-23, EmaA signal peptide with amino acids 2 to 23 deleted; EmaASPΔ24-53, EmaA signal peptide with amino acids 24 to 53 deleted; EmaASPΔ2-35, EmaA signal peptide with amino acids 2 to 35 deleted; EmaASPΔ16-39, EmaA signal peptide with amino acids 16 to 39 deleted. (B) Immunoblot analysis of alkaline phosphatase expression. Whole-cell lysates, derived from the E. coli CC118 strains expressing the PhoA constructs described above for panel A, were probed with anti-PhoA antiserum. The constructs are shown in the same order as in panel A.
The EmaA signal peptide drives secretion of the epithelial cell adhesin Aae in A. actinomycetemcomitans.
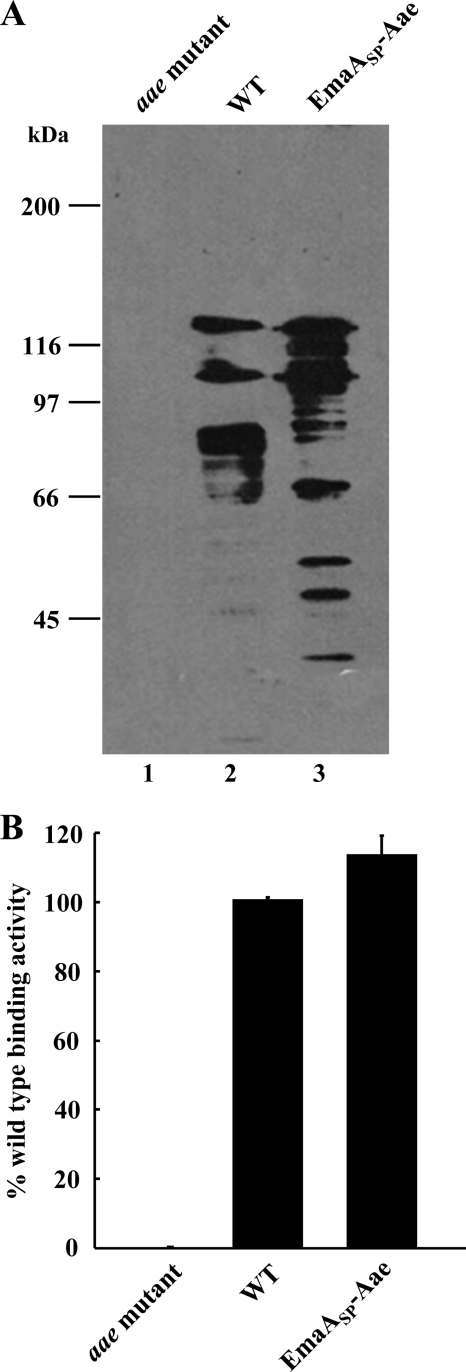
Based on these alkaline phosphatase studies, we generated a fusion construct to determine whether the first 56 amino acids of the EmaA protein sequence also function as a signal peptide in A. actinomycetemcomitans. The predicted EmaA signal peptide sequence was fused in-frame with the passenger domain of the typical A. actinomycetemcomitans autotransporter Aae, a known epithelial cell adhesin (41), and transformed into an aae mutant strain. Immunoblot analysis of membrane proteins from the strain containing this construct (EmaASP-Aae) demonstrated the presence of immunoreactive material associated with the membrane fraction (Fig. 3A, lane 3). The staining pattern is comparable to that of the A. actinomycetemcomitans wild-type membrane fraction (Fig. 3A, lane 2). Immunoreactivity was not associated with membrane proteins derived from the aae mutant strain (Fig. 3A, lane 1). The immunoreactive bands with a molecular mass lower than that of the intact protein (130 kDa) (Fig. 3A, lanes 2 and 3) are proposed to be degradation products of Aae.
Fig. 3.
Membrane and surface localization of the A. actinomycetemcomitans epithelial cell adhesin Aae. (A) Immunoblot analysis of bacterial membranes probed with a polyclonal antiserum specific for Aae. Lane 1, aae mutant strain; lane 2, wild-type (WT) strain (VT1169); lane 3, aae mutant strain transformed with a replicating plasmid expressing the EmaA signal peptide-Aae chimera (EmaASP-Aae). (B) Detection of Aae on the bacterial surface. Surface-exposed Aae was detected using antibodies specific for Aae in an ELISA format with bacteria immobilized on the bottom of wells on 96-well microtiter plates.
In the strain expressing the Aae chimera, Aae was localized to the bacterial surface, as indicated in Fig. 3B. Whole bacteria immobilized onto a solid surface and probed with anti-Aae serum demonstrated a statistically significant difference in antibody binding activity between the strain expressing the Aae chimera and the aae mutant strain. A strain expressing wild-type levels of Aae was used as the positive control for surface expression of Aae. The Aae surface expression levels were equivalent in both the wild-type strain and the Aae chimera-expressing strains (Fig. 3B).
The N-terminal extension and canonical signal peptide regions are required for protein secretion.
The EmaA long signal peptide can be divided into the N-terminal extension and the canonical N, H, C region (Fig. 1B). Therefore, deletion mutants of the N-terminal extension (amino acids 2 to 23) and the canonical region (amino acids 24 to 56) were constructed and investigated for membrane translocation. In E. coli, the strain expressing EmaASPΔ2-23 (EmaASP with amino acids 2 to 23 deleted) displayed PhoA activity but at levels much lower than the full-length sequence (Fig. 2A). This is in contrast to the strain expressing EmaASPΔ24-53 (EmaASP with amino acids 24 to 53 deleted) that lacked PhoA activity (Fig. 2A). The lack of enzyme activity correlated with the absence of PhoA protein as determined by immunoblot analysis in E. coli (Fig. 2B).
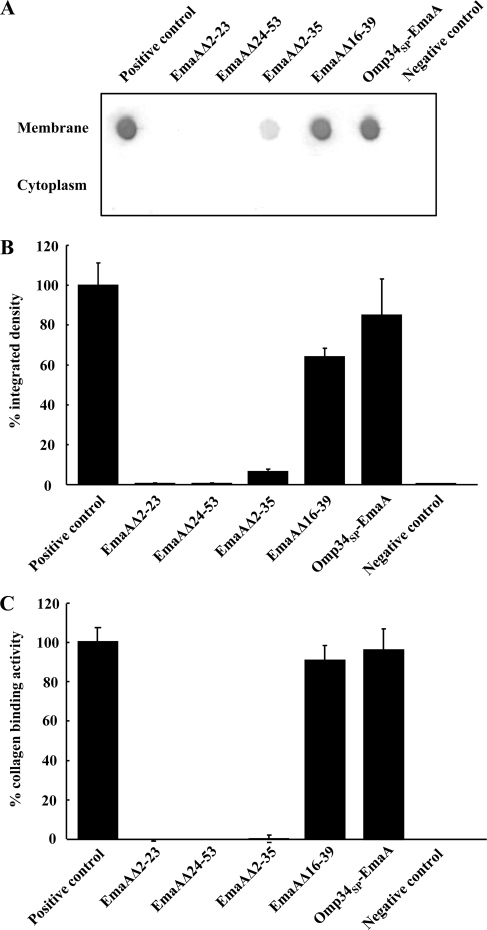
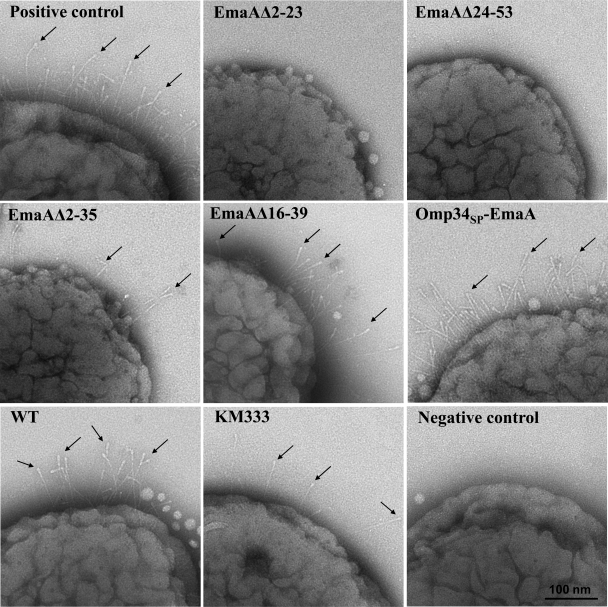
A. actinomycetemcomitans strains transformed with these signal peptide mutations in the emaA gene were assessed for EmaA localization in the cytoplasm or membrane fractions using an anti-EmaA monoclonal antibody (58). EmaA forms aggregates upon solubilization and heating, which does not allow for reproducible entry and separation of the monomers in SDS-polyacrylamide gels (58). Immunoreactive EmaA is usually associated with the wells of the stacking gel, and only a small amount of monomer is present at the expected molecular weight. Therefore, for a more reliable quantification of the amount of EmaA produced by the strains used in this study, an immunodot blot format was used. Fractionation of the wild-type bacterial cells into cytoplasm and membrane fractions indicated that EmaA was localized to the membrane fraction, with little, if any, immunoreactive protein present in the cytoplasm at the concentrations used (Fig. 4A). In A. actinomycetemcomitans, EmaA was not detected in the cytoplasm or the membrane fraction of the strains expressing either EmaAΔ2-23 or EmaAΔ24-53 sequences (Fig. 4B). In addition, EmaA structures were absent on the surfaces of these strains (Fig. 5). Quantitative real-time PCR (QRT-PCR) analysis of these constructs suggested the absence of any defect in transcription of emaA mRNA compared with the fully complemented strain (data not shown). Associated with the absence of structures was the loss of collagen binding activity of these strains, which was similar to the emaA mutant strain containing the empty vector (Fig. 4C). The data indicated that neither of the amino acid sequences 2-23 or 24-53 of the signal peptide can support EmaA secretion alone.
Fig. 4.
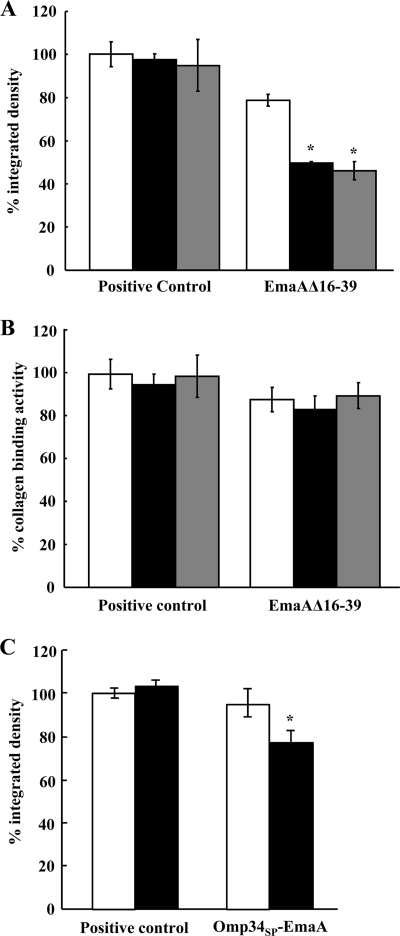
Immunodot blot analysis and collagen binding activity of A. actinomycetemcomitans strains expressing the intact and altered EmaA signal peptides. (A) Immunodot blot assay for EmaA from membrane and cytoplasm fractions. Membrane and cytoplasm fractions were generated from the emaA null mutant transformed with individual plasmids containing sequence encoding either the native long signal peptide or various portions of the long signal peptide. Positive control, emaA mutant strain transformed with a plasmid encoding the intact EmaA signal peptide; EmaAΔ2-23, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 2 to 23; EmaAΔ24-53, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 24 to 53; EmaAΔ2-35, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 2 to 35; EmaAΔ16-39, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 16 to 39; Omp34SP-EmaA, emaA mutant strain transformed with a plasmid encoding the substitution of the EmaA signal peptide with the Omp34 signal peptide; negative control, emaA mutant strain transformed with the empty plasmid. (B) Relative integrated densities of the immunodot blot of EmaA expressed by the strains containing the constructs described above for panel A. (C) Collagen binding activities of the strains in panels A and B as measured by ELISA.
Fig. 5.
Whole-mount transmission electron micrographs of A. actinomycetemcomitans strains negatively stained with Nano-W. Positive control, emaA mutant strain transformed with a plasmid encoding the intact EmaA signal peptide; EmaAΔ2-23, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 2 to 23; EmaAΔ24-53, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 24 to 53; EmaAΔ2-35, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 2 to 35; EmaAΔ16-39, emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with deletion of amino acids 16 to 39; Omp34SP-EmaA, emaA mutant strain transformed with a plasmid encoding the Omp34 signal peptide instead of the EmaA signal peptide; WT, wild-type strain transformed with empty plasmid; KM333, chromosomal deletion strain of EmaA amino acids 16 to 39; negative control, emaA mutant strain transformed with the empty plasmid. The small black arrows indicate EmaA structures on the bacterial surface.
Deletion of a portion of the canonical signal peptide region allows for surface expression but abolished collagen binding activity.
An in-frame EmaA signal peptide deletion construct, corresponding to deletion of amino acids 2 to 35, was generated and assayed for PhoA activity in E. coli. The strain expressing this construct (EmaASPΔ2-35) demonstrated marginal PhoA activity, compared with the intact signal peptide (Fig. 2A). In A. actinomycetemcomitans, the strain expressing the plasmid with this deletion in the emaA gene (EmaAΔ2-35) contained less than 10% of the EmaA protein associated with the membrane fraction compared with the strain expressing the full-length signal peptide (Fig. 4B). EmaA in the cytoplasm of this strain was not detected (Fig. 4A). Transcriptional activity of the EmaAΔ2-35 plasmid was similar to the plasmid containing the full-length signal peptide (data not shown).
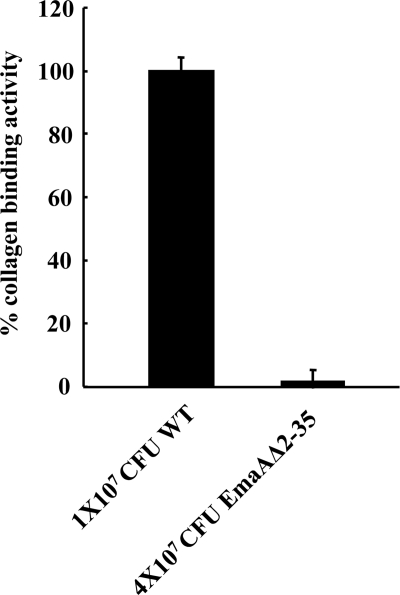
Transmission electron microscopy images indicate the presence of EmaA structures on the surface of this strain (Fig. 5). However, the reduced frequency of the visualization of EmaA structures in these images suggested that the number of EmaA structures on the surface of this strain was greatly diminished compared with the positive-control strain. Associated with the reduction of EmaA structures was the loss of collagen binding activity in the EmaAΔ2-35-expressing strain (Fig. 4C). The binding activity of the EmaAΔ2-35-expressing strain was similar to the binding activity of the emaA mutant strain, which does not synthesize any detectable EmaA protein or show any structures on the bacterial surface (Fig. 4C and 5). Since the collagen binding activity of these strains was determined using equal cell numbers in the ELISA, we posited that the difference in collagen binding activity could be attributed to the amount of EmaA synthesized. Therefore, the number of cells of the EmaAΔ2-35-expressing strain and wild-type strain added to the assay was first normalized for EmaA protein, as determined by immunodot blot analysis (4 × 107 and 1 × 107 CFU, respectively). In this series of experiments, significantly more collagen binding activity was associated with the cells of the wild-type strain compared with the activity generated by four times the number of EmaAΔ2-35-expressing cells (Fig. 6). The lack of collagen binding activity of the EmaAΔ2-35-expressing strain does not appear to be attributed to the absence of EmaA structures on the cell surface.
Fig. 6.
Collagen binding activities of A. actinomycetemcomitans strains. Collagen binding activity of 1 × 107 CFU WT (wild-type strain) or 4 × 107 CFU EmaAΔ2-35 (emaA mutant strain transformed with pKMΔ2-35) as measured by ELISA.
The full-length EmaA signal peptide is not absolutely required for protein secretion.
The PhoA activity of a strain expressing a chimera of the EmaA signal peptide with deletion of amino acids 16 to 39 (EmaASPΔ16-39) fused to PhoA in E. coli was similar to the activity of the strain expressing the signal peptide construct with deletion of amino acids 2 to 23 (EmaASPΔ2-23) (Fig. 2). In A. actinomycetemcomitans, the EmaAΔ2-23-expressing strain did not target EmaA to the membrane. Interestingly, the strain with the deletion of amino acids 16 to 39 of the EmaA signal peptide targeted the protein to the membrane in A. actinomycetemcomitans (Fig. 4B). The electron micrographs indicated that structures were present on the surface and suggest that the proteins are competent for outer membrane secretion (Fig. 5). The change in the composition of the signal peptide did not result in increased EmaA protein aggregation, which may coisolate with the membrane fraction (data not shown).
Quantification of the EmaA present in the membrane fraction of the EmaAΔ16-39-expressing strain demonstrated a significant decrease (∼30%) in the amount of EmaA compared with the strain transformed with the full-length sequence. However, the decrease in the amount of EmaA in the membrane did not affect the collagen binding activity of this strain compared with the full-length emaA transformant (Fig. 4C). This suggests that amino acids 16 to 39 of the signal peptide facilitate but are not absolutely necessary for EmaA secretion.
Mutation of amino acids 16 to 39 of the EmaA signal peptide impairs protein localization at elevated temperatures.
The amount of EmaA produced by strains expressing the mutant signal peptide EmaAΔ16-39 and the full-length signal peptide grown at different temperatures was investigated. When grown at either 37°C or 39°C, the amounts of EmaA synthesized by the strain transformed with full-length signal peptide were similar (Fig. 7A). However, there was a statistically significant decrease (P < 0.01) in the amount of EmaA synthesized by the EmaAΔ16-39-expressing strain grown at 39°C compared to the strain grown at 37°C. Similar results were seen when the strain was heat shocked at 42°C (Fig. 7A). Both strains displayed EmaA structures on the surfaces of the bacteria (Fig. 5) and were competent to form active structures (Fig. 7B). However, the change in the amount of EmaA in the EmaAΔ16-39-expressing strain was not reflected in the collagen binding activity. The collagen binding activity was equivalent for this strain and the full-length transformed strain at the different temperatures (Fig. 7B).
Fig. 7.
EmaA immunodot blot and collagen binding activities of transformed A. actinomycetemcomitans strains. (A) Bacterial lysates (corresponding to 4 × 107 CFU bacteria) were prepared and immobilized on nitrocellulose membranes. EmaA was detected using a monoclonal antibody specific for EmaA in cells grown at 37°C (black bars) or 39°C (white bars) or heat shocked at 42°C (gray bars). Positive control, emaA mutant strain expressing the intact EmaA protein (the strain was transformed with pKM9); EmaAΔ16-39, emaA mutant strain expressing the EmaA protein with amino acids 16 to 39 deleted (the strain was transformed with pKMΔ16-39). (B) Collagen binding activity of the strain expressing intact EmaA (positive control) or strain EmaAΔ16-39 (emaA mutant strain transformed with a plasmid encoding the EmaA signal peptide with amino acids 16 to 39 deleted) grown at 37°C (black bars) or 39°C (white bars) or heat shocked at 42°C (gray bars) as measured by ELISA. (C) Integrated intensities of the immunoreactive protein in the membrane of cells grown at 37°C (white bars) or heat shocked at 42°C (black bars). Positive control, emaA mutant strain expressing the intact EmaA protein (the strain was transformed with pKM9); Omp34SP-EmaA, emaA mutant strain transformed with a plasmid encoding the Omp34 signal peptide instead of the EmaA signal peptide.
The lack of a difference in collagen binding activity between the full-length and the EmaAΔ16-39-expressing strains was considered to be attributed to the number of EmaA structures on the surfaces of the bacteria (Fig. 5). Overexpression of the protein may mask small changes in the collagen binding activity of this strain due to saturation binding kinetics, attributed to the copy number of the plasmid. Hence, we generated and characterized a strain (KM333) containing an in-frame chromosomal deletion of amino acids 16 to 39 of the EmaA signal peptide to minimize the number of copies of emaA.
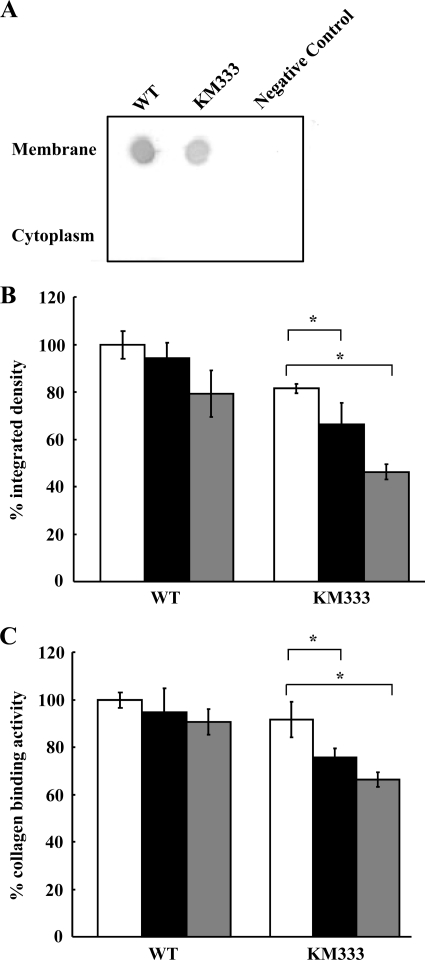
A decrease in the amount of EmaA synthesized was observed in the chromosomal deletion strain (KM333); this was similar to the decrease found associated with the transformant expressing the multicopy plasmid (Fig. 8A). The electron micrographs suggest that EmaA structures are present on the surface of the chromosomal deletion strain but in reduced numbers compared with the plasmid-expressing strain (EmaAΔ16-39) (Fig. 5). The KM333 strain, either adapted to growth at 39°C or heat shocked at 42°C, demonstrated a decreased collagen binding activity (Fig. 8C). This is in contrast to the lack of a difference in collagen binding activity of the strain expressing the multicopy plasmid. There was no difference in collagen binding activity of the wild-type strain when grown at the different temperatures, suggesting a role for this sequence of the signal peptide in the expression of EmaA at elevated temperatures.
Fig. 8.
EmaA expression and collagen binding activities of Δ16-39 (amino acids 16 to 39 deleted) signal peptide chromosomal deletion strain (KM333) and wild-type strain. (A) Immunodot blot assay of membrane and cytoplasm fractions from WT (wild-type strain), KM333 (Δ16-39 signal peptide chromosomal deletion strain), and negative control (emaA mutant strain) grown at 37°C. (B) Bacterial lysates (corresponding to 4 × 107 CFU bacteria) were prepared and immobilized on nitrocellulose membranes. EmaA was detected using a monoclonal antibody specific for EmaA. Relative integrated densities of EmaA expression from the WT (wild-type strain) or KM333 (Δ16-39 signal peptide chromosomal deletion strain) grown at 37°C (white bars) or 39°C (black bars) or heat shocked at 42°C for 3 h (gray bars). (C) Collagen binding activity of WT (wild type) or KM333 (Δ16-39 signal peptide chromosomal deletion strain) grown at 37°C (white bars) or 39°C (black bars), or heat shocked at 42°C for 3 h (gray bars) as measured by ELISA. Values that are statistically significantly different (P < 0.05) are indicated by the brackets and an asterisk.
A typical signal peptide is sufficient for EmaA secretion.
A fusion construct composed of the signal peptide of Omp34 (55) and the passenger domain of EmaA (Omp34SP-EmaA) was generated to determine whether a typical signal peptide (N, H, C region) would support EmaA protein secretion. In the emaA mutant strain transformed with the Omp34SP-EmaA fusion construct, EmaA was located in the membrane fraction (Fig. 4A and B) in amounts similar to the strain expressing the complete emaA gene.
Electron micrographs of the Omp34SP-EmaA-expressing strain clearly demonstrated the ability of the translocated proteins to form EmaA antenna-like structures on the bacterial surface (Fig. 5). This is in stark contrast to the surface of the emaA mutant strain, which does not display any EmaA surface structures (Fig. 5, negative control). The proper oligomerization of the EmaA monomers to form a functionally active structure, in terms of collagen binding, was also determined for these strains. The collagen binding activity of the Omp34SP-EmaA-expressing strain was similar to the emaA mutant strain transformed with the entire emaA gene on the same plasmid backbone (Fig. 4C).
The long signal peptide of EmaA was required for maximum secretion at elevated temperatures (Fig. 7). To determine if a typical signal peptide responds to elevated temperatures similar to the long signal peptide, the Omp34SP-EmaA-expressing strain was grown under heat shock conditions and analyzed for the amount of EmaA present in the membrane. As demonstrated in Fig. 7C, there was a statistically significant decrease in the amount of membrane-localized EmaA in the heat-shocked strain compared with the same strain grown at 37°C.
DISCUSSION
The collagen-binding adhesin of A. actinomycetemcomitans, EmaA, is a trimeric autotransporter, which contains a long signal peptide as demonstrated by this study. The results of deletion analysis, in the context of PhoA activity in E. coli, suggest that the entire amino acid sequence is necessary for full translocation activity. The membrane and surface localization of the EmaASP-Aae fusion protein in A. actinomycetemcomitans strongly suggests that the sequence made up of the first 56 amino acids of the EmaA protein provides the molecular signal(s) required for inner membrane targeting, translocation, surface expression, and proper functional activity under physiological conditions.
The N-terminal extension or extended signal peptide region is suggested to be involved in the delay of protein translocation for some proteins containing long peptides (13, 37, 47). However, in other systems, this sequence has no effect on translocation (21). In our study, the deletion of the region of protein corresponding to the N-terminal extension, using PhoA as the reporter, resulted in reduced translocation. Furthermore, the strain expressing the canonical region of the EmaA peptide fused with PhoA did not demonstrate any enzymatic activity. Together, these data suggest that the N-terminal extension of EmaA, in this heterologous system, alone has no activity but enhances the canonical signal peptide activity.
In A. actinomycetemcomitans, the strain expressing only the canonical region of the EmaA peptide fused to EmaA was not competent for EmaA membrane localization compared with the activity of the same sequence fused to PhoA in E. coli. This apparent contradiction in the two systems may be attributed to the difference in the amino acid sequence of the protein adjacent to the signal peptide (PhoA versus EmaA). Studies suggest that the protein sequence adjacent to the signal peptide is important for the interaction with the protein targeting and translocation machinery (1, 22). EmaA was also not observed in the membrane of the strain expressing only the N-terminal extension fused to EmaA. However, both strains were shown to express similar levels of emaA mRNA, which argues against a defect in transcription of the plasmids. Furthermore, the addition or deletion of extra amino acids to the sequences (based on the deduced protein sequence) did not affect the outcome of these experiments (data not shown). Hence, the data indicate that the full-length signal sequence may be important for the stability of the newly synthesized polypeptide chain preventing the degradation of the translated protein, as ascribed for one of the functions of the signal peptide (51).
In other bacterial systems, the canonical sequences are functional for protein membrane targeting and translocation (21, 37, 47). The membrane translocation activity of this region of the signal peptide of the secreted hemoglobin protease, Hbp, from E. coli is identical to the activity of the full-length signal peptide, which indicates that the N-terminal extension is not required for activity (21). The canonical region of the signal peptide of the monomeric autotransporter serine protease of E. coli, EspP, is also functional for membrane targeting and translocation, although at a reduced level compared to the full-length signal peptide (37, 47). The corresponding regions of the signal peptide of the trimeric autotransporter EmaA do not support membrane targeting and translocation, suggesting a difference in the function of the long signal peptide of EmaA in A. actinomycetemcomitans. Studies of the signal peptide of other members of the trimeric autotransporter family have not been reported.
The N-terminal extension and the canonical region of the long signal peptide of EmaA are not sufficient for membrane localization of EmaA in A. actinomycetemcomitans. However, a sequence within the canonical region (amino acids 36 to 56) demonstrated activity in both bacterial systems, albeit at reduced levels. The presence of EmaA structures and the absence of collagen binding activity associated with this mutant suggest that this sequence allows for normal EmaA localization but abolishes the function of the adhesin. Studies propose that the secondary structure of the signal peptide modulates the cleavage specificity of the signal peptidase (20, 38). Therefore, a change in the secondary structure of the EmaAΔ2-35 signal peptide may alter the peptidase cleavage site, resulting in modified EmaA monomers that are not competent to fold into an active conformation (58). Alternatively, the lack of collagen binding of this strain may be due to a reduction in the number of EmaA surface structures per cell.
Inspection of the long signal peptide sequence of EmaA suggested that deletion of amino acids 16 to 39 of the full-length sequence would result in a peptide that resembles a typical signal peptide (Fig. 1B). Our data clearly demonstrate that in this strain, EmaA is transported to the membrane and displays surface structures that interact with collagen. The data suggest that amino acids 16 to 39 facilitate maximal secretion but are not completely essential for protein targeting and translocation. Furthermore, the data indicate that amino acids 40 to 53, in context with the N-terminal extension, are important or required for the interaction of the peptide with the protein secretion machinery.
A distinct role for long signal peptides has not yet been addressed in other organisms. We hypothesize that the additional amino acids (amino acids 16 to 39) in the EmaA signal peptide function to ensure the secretion of this adhesin under physiological stress. These stresses may include changes in the environment of the gingival sulcus, which occurs during the transition between health and disease (2, 35, 43, 44). These conditions may contribute to a stress response in the bacterium, which translates to an overall change in the transcriptional activity of specific gene products, including heat shock proteins or stress-induced proteins (56). These molecular chaperones modulate polypeptide folding, assembly, degradation, and translocation (9, 24, 25). We observed a reduction in the amount of EmaA and collagen binding of the chromosomal EmaAΔ16-39 mutant strain (KM333) grown at increased temperatures compared with the same strain grown at 37°C. This difference in the amount of EmaA protein and collagen binding activity was not observed with the wild-type strain grown under identical conditions. Interestingly, we have demonstrated that a typical signal peptide can drive secretion of EmaA. However, in this strain, the reduction in the amount of EmaA secreted at 42°C (Fig. 7C) supports the hypothesis that amino acids 16 to 39 of the long signal peptide are important, in part, for the proper protein folding of this adhesin at elevated temperatures. The experimental data using the typical signal peptide of Omp34 further corroborate our hypothesis.
The binding of A. actinomycetemcomitans to collagen, mediated by EmaA, is important for the initiation of infective endocarditis (48) and may contribute to the pathogenicity of this bacterium in periodontal diseases. EmaA is the only protein that contains an extended signal peptide of the membrane proteins in A. actinomycetemcomitans that have been characterized, such as Aae, Omp34, and Omp100 (23, 41, 55). The data presented in this study suggest that the amino acids of the extended signal peptide of EmaA are required for protein stability in the cytoplasm and the proper assembly of the monomers to form a functional adhesin. Furthermore, specific amino acids of the long signal peptide may be important for the presentation of this adhesin on the bacterial surface to bind and colonize the oral cavity under changing environmental conditions.
ACKNOWLEDGMENTS
We thank Chris Lennox for his contribution to this project.
This research was supported by National Institutes of Health-National Institute of Dental and Craniofacial Research (NIH-NIDCR) grant RO1-DE13824.
Footnotes
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Andrews D. W., Perara E., Lesser C., Lingappa V. R. 1988. Sequences beyond the cleavage site influence signal peptide function. J. Biol. Chem. 263: 15791–15798 [PubMed] [Google Scholar]
- 1a. Applied Biosystems 1997. ABI PRISM 7700 sequence detection system: user bulletin #2. Relative quantification of gene expression. Applied Biosystems, Foster City, CA [Google Scholar]
- 2. Baab D. A., Oberg A., Lundstrom A. 1990. Gingival blood flow and temperature changes in young humans with a history of periodontitis. Arch. Oral Biol. 35: 95–101 [DOI] [PubMed] [Google Scholar]
- 3. Belin D., Bost S., Vassalli J. D., Strub K. 1996. A two-step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 15: 468–478 [PMC free article] [PubMed] [Google Scholar]
- 4. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- 5. Brickman E., Beckwith J. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96: 307–316 [DOI] [PubMed] [Google Scholar]
- 6. Carlile J. R., Beckman E. N., Arensman R. M. 1984. Actinobacillus actinomycetemcomitans pneumonia. Clin. Pediatr. (Phila.) 23: 578–580 [DOI] [PubMed] [Google Scholar]
- 7. Chenna R., et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christersson L. A. 1993. Actinobacillus actinomycetemcomitans and localized juvenile periodontitis. Clinical, microbiologic and histologic studies. Swed. Dent. J. Suppl. 90: 1–46 [PubMed] [Google Scholar]
- 9. Collier D. N., Bankaitis V. A., Weiss J. B., Bassford P. J., Jr 1988. The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell 53: 273–283 [DOI] [PubMed] [Google Scholar]
- 10. Dautin N., Bernstein H. D. 2007. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 61: 89–112 [DOI] [PubMed] [Google Scholar]
- 11. Desvaux M., et al. 2006. The unusual extended signal peptide region of the type V secretion system is phylogenetically restricted. FEMS Microbiol. Lett. 264: 22–30 [DOI] [PubMed] [Google Scholar]
- 12. Desvaux M., Parham N. J., Henderson I. R. 2004. Type V protein secretion: simplicity gone awry? Curr. Issues Mol. Biol. 6: 111–124 [PubMed] [Google Scholar]
- 13. Desvaux M., et al. 2007. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology 153: 59–70 [DOI] [PubMed] [Google Scholar]
- 14. Gallant C. V., Sedic M., Chicoine E. A., Ruiz T., Mintz K. P. 2008. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190: 5972–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grant S. G., Jessee J., Bloom F. R., Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87: 4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hegde R. S., Bernstein H. D. 2006. The surprising complexity of signal sequences. Trends Biochem. Sci. 31: 563–571 [DOI] [PubMed] [Google Scholar]
- 17. Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68: 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrero M., de Lorenzo V., Timmis K. N. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172: 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacob-Dubuisson F., Locht C., Antoine R. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40: 306–313 [DOI] [PubMed] [Google Scholar]
- 20. Jain R. G., Rusch S. L., Kendall D. A. 1994. Signal peptide cleavage regions. Functional limits on length and topological implications. J. Biol. Chem. 269: 16305–16310 [PubMed] [Google Scholar]
- 21. Jong W. S., Luirink J. 2008. The conserved extension of the Hbp autotransporter signal peptide does not determine targeting pathway specificity. Biochem. Biophys. Res. Commun. 368: 522–527 [DOI] [PubMed] [Google Scholar]
- 22. Kajava A. V., Zolov S. N., Kalinin A. E., Nesmeyanova M. A. 2000. The net charge of the first 18 residues of the mature sequence affects protein translocation across the cytoplasmic membrane of gram-negative bacteria. J. Bacteriol. 182: 2163–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komatsuzawa H., et al. 2002. Identification of six major outer membrane proteins from Actinobacillus actinomycetemcomitans. Gene 288: 195–201 [DOI] [PubMed] [Google Scholar]
- 24. Kumamoto C. A., Beckwith J. 1985. Evidence for specificity at an early step in protein export in Escherichia coli. J. Bacteriol. 163: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumamoto C. A., Gannon P. M. 1988. Effects of Escherichia coli secB mutations on pre-maltose binding protein conformation and export kinetics. J. Biol. Chem. 263: 11554–11558 [PubMed] [Google Scholar]
- 26. Kurys G., Tagaya Y., Bamford R., Hanover J. A., Waldmann T. A. 2000. The long signal peptide isoform and its alternative processing direct the intracellular trafficking of interleukin-15. J. Biol. Chem. 275: 30653–30659 [DOI] [PubMed] [Google Scholar]
- 27. Mauff A. C., Miller S., Kuhnle V., Carmichael M. 1983. Infections due to Actinobacillus actinomycetemcomitans. A report of 3 cases. S. Afr. Med. J. 63: 580–581 [PubMed] [Google Scholar]
- 28. Mintz K. P. 2004. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 150: 2677–2688 [DOI] [PubMed] [Google Scholar]
- 29. Mintz K. P., Brissette C., Fives-Taylor P. M. 2002. A recombinase A-deficient strain of Actinobacillus actinomycetemcomitans constructed by insertional mutagenesis using a mobilizable plasmid. FEMS Microbiol. Lett. 206: 87–92 [DOI] [PubMed] [Google Scholar]
- 30. Mintz K. P., Fives-Taylor P. M. 1994. Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect. Immun. 62: 3672–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mintz K. P., Fives-Taylor P. M. 1999. Binding of the periodontal pathogen Actinobacillus actinomycetemcomitans to extracellular matrix proteins. Oral Microbiol. Immunol. 14: 109–116 [DOI] [PubMed] [Google Scholar]
- 32. Muhle I., Rau J., Ruskin J. 1979. Vertebral osteomyelitis due to Actinobacillus actinomycetemcomitans. JAMA 241: 1824–1825 [PubMed] [Google Scholar]
- 33. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6 [DOI] [PubMed] [Google Scholar]
- 34. Nielsen H., Krogh A. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 122–130 [PubMed] [Google Scholar]
- 35. Page R. C., Schroeder H. E. 1976. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab. Invest. 34: 235–249 [PubMed] [Google Scholar]
- 36. Pearce B. J., Yin Y. B., Masure H. R. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9: 1037–1050 [DOI] [PubMed] [Google Scholar]
- 37. Peterson J. H., Szabady R. L., Bernstein H. D. 2006. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J. Biol. Chem. 281: 9038–9048 [DOI] [PubMed] [Google Scholar]
- 38. Pratap J., Dikshit K. L. 1998. Effect of signal peptide changes on the extracellular processing of streptokinase from Escherichia coli: requirement for secondary structure at the cleavage junction. Mol. Gen. Genet. 258: 326–333 [DOI] [PubMed] [Google Scholar]
- 39. Priefer U. B., Simon R., Puhler A. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163: 324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reider J., Wheat J. 1979. Endocarditis caused by Actinobacillus actinomycetemcomitans. South. Med. J. 72: 1219–1220 [DOI] [PubMed] [Google Scholar]
- 41. Rose J. E., Meyer D. H., Fives-Taylor P. M. 2003. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 71: 2384–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruiz T., Lenox C., Radermacher M., Mintz K. P. 2006. Novel surface structures are associated with the adhesion of Actinobacillus actinomycetemcomitans to collagen. Infect. Immun. 74: 6163–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Slots J., Listgarten M. A. 1988. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J. Clin. Periodontol. 15: 85–93 [DOI] [PubMed] [Google Scholar]
- 44. Slots J., Reynolds H. S., Genco R. J. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29: 1013–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sreenivasan P. K., LeBlanc D. J., Lee L. N., Fives-Taylor P. 1991. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect. Immun. 59: 4621–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. St. Geme J. W., III, Cutter D. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182: 6005–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szabady R. L., Peterson J. H., Skillman K. M., Bernstein H. D. 2005. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc. Natl. Acad. Sci. U. S. A. 102: 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang G., Kitten T., Munro C. L., Wellman G. C., Mintz K. P. 2008. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect. Immun. 76: 2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang G., Mintz K. P. 2010. Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway. J. Bacteriol. 192: 1395–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40: 397–413 [DOI] [PubMed] [Google Scholar]
- 51. von Heijne G. 1990. The signal peptide. J. Membr. Biol. 115: 195–201 [DOI] [PubMed] [Google Scholar]
- 52. von Heijne G. 1985. Signal sequences. The limits of variation. J. Mol. Biol. 184: 99–105 [DOI] [PubMed] [Google Scholar]
- 53. von Heijne G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2: 531–534 [DOI] [PubMed] [Google Scholar]
- 54. Westerlund B., Korhonen T. K. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9: 687–694 [DOI] [PubMed] [Google Scholar]
- 55. White P. A., Nair S. P., Kim M. J., Wilson M., Henderson B. 1998. Molecular characterization of an outer membrane protein of Actinobacillus actinomycetemcomitans belonging to the OmpA family. Infect. Immun. 66: 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamamori T., Yura T. 1980. Temperature-induced synthesis of specific proteins in Escherichia coli: evidence for transcriptional control. J. Bacteriol. 142: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu C., Mintz K. P., Ruiz T. 2009. Investigation of the three-dimensional architecture of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans by electron tomography. J. Bacteriol. 191: 6253–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu C., Ruiz T., Lenox C., Mintz K. P. 2008. Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zambon J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12: 1–20 [DOI] [PubMed] [Google Scholar]