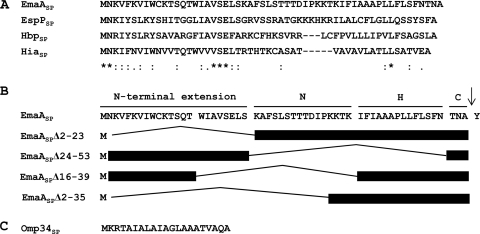
Fig. 1.
Signal peptides of selected Gram-negative bacterial proteins. (A) Sequence alignment of long signal peptides of proteins from different organisms. Alignments were generated using ClustalW2 (7). Identical amino acids (*), highly conserved amino acids (:), and weakly conserved amino acids (.) are indicated below the sequence alignment. Gaps introduced to maximize sequence alignment are indicated by the dashes. (B) Predicted domain structure of the EmaA signal peptide and schematics of the in-frame EmaA signal peptide deletion constructs used in this study. The arrow indicates the predicted signal peptidase cleavage site between amino acid 56 (Ala) and 57 (Tyr). (C) Sequence of the typical signal peptide of Omp34 of A. actinomycetemcomitans.