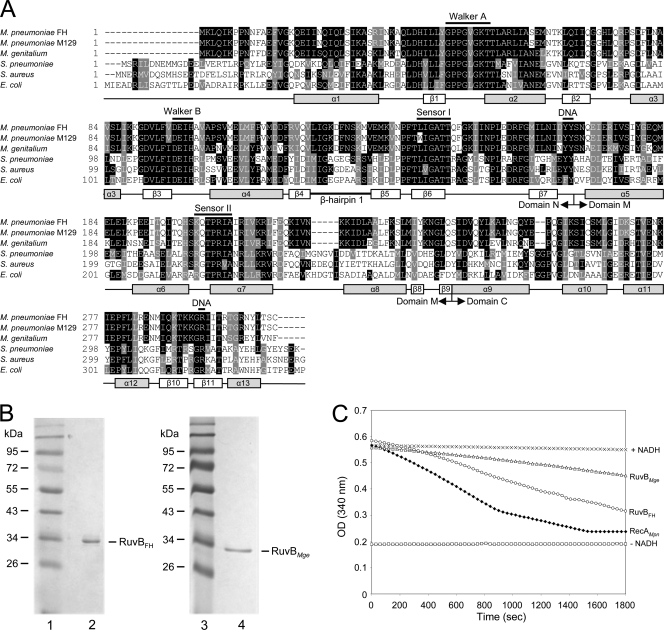
Fig. 1.
Multiple alignment of RuvB sequences and ATPase activity of RuvBFH and RuvBMge. (A) A multiple alignment was generated with amino acid sequences predicted to be encoded by the following ORFs (with GenBank accession numbers in parentheses): M. pneumoniae FH MPN536 (ADK87167), M. pneumoniae M129 MPN536 (AAB95954), M. genitalium G37 MG359 (ZP_05405689), Streptococcus pneumoniae ruvB (Q97SR6), Staphylococcus aureus ruvB (NP_374754), and E. coli ruvB (P0A812). Predicted secondary structural features of the RuvB proteins are shown below the alignment and are based on the crystal structures of the RuvB proteins from Thermus thermophilus HB8 (45) and Thermotoga maritima (28). The annotation of the (predicted) α helices and β strands, as well as the boundaries of the amino-terminal (N), central (M), and carboxyl-terminal domains, is derived from Yamada et al. (45). The positions of crucial, conserved motifs of AAA+ proteins, i.e., Walker A, Walker B, and sensor I and sensor II motifs, are indicated above the sequences. Amino acids potentially involved in DNA or nucleotide binding are also indicated (DNA) (12, 45).The multiple alignment was performed using Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The program BOXSHADE, version 3.21 (http://www.ch.embnet.org/software/BOX_form.html), was used to generate white letters on black boxes (for residues that are identical in at least three out of six sequences) and white letters on gray boxes (for similar residues). (B) Purification of RuvBFH and RuvBMge. Samples of purified RuvBFH (left panel, lane 2) and RuvBMge (right panel, lane 4) were analyzed by SDS-PAGE (12%) and Coomassie brilliant blue (CBB) staining. The sizes of protein markers (lanes 1 and 3; PageRuler Prestained Protein Ladder [Fermentas]) are shown on the left-hand side of each panel in kDa. (C) ATPase activity of RuvBFH and RuvBMge. ATP hydrolysis by RuvBFH (○) and RuvBMge (Δ) was measured at a protein concentration of 0.5 μM in the presence of Mg2+ (1 mM) and ϕX174 single-stranded DNA (1.5 nM). The ATPase activity was determined using an NADH-coupled assay. Using this assay, the activity is calculated from the stationary velocities of ATP hydrolysis as determined by monitoring the absorption of NADH at 340 nm (19, 34). The RecAMpn protein, characterized previously by Sluijter et al. (34), was taken along as a positive control at a concentration of 0.5 μM (♦). Control reactions were performed in the absence of any protein (+NADH, ×) and in the absence of both NADH and protein (−NADH, □). OD, optical density.