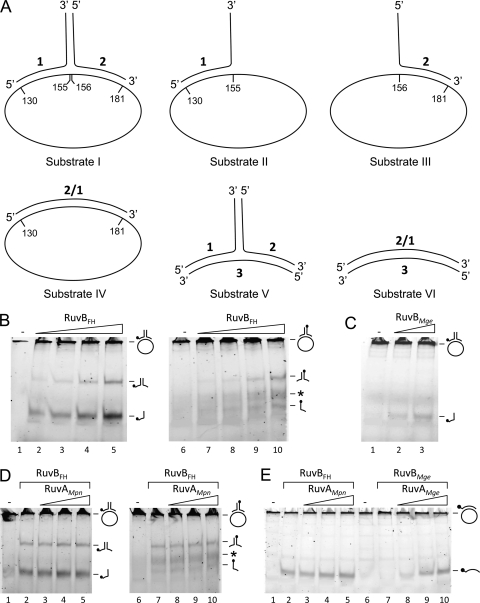
Fig. 3.
DNA helicase activity of RuvBFH and RuvBMge. (A) Schematic illustration of the DNA substrates that were used in the DNA helicase assays. The substrates are composed of oligonucleotides (substrates V and VI) or a combination of oligonucleotides and single-stranded, circular 5,386-bp ϕX174 DNA (substrates I to IV). The ϕX174 DNA is pictured as an ellipse and is not drawn to scale with respect to the oligonucleotides. The sequences of the oligonucleotides (oligonucleotides 1, 2, 2/1, and 3) were described by Tsaneva et al. (40). (B) The activity of RuvBFH on substrate I. Substrate I, 6-FAM labeled at the 5′ end of either oligonucleotide 1 (lanes 1 to 5) or oligonucleotide 2 (lanes 6 to 10), was incubated with either 0 μM (−; lanes 1 and 6), 0.1 μM (lanes 2 and 7), 0.3 μM (lanes 3 and 8), 0.9 μM (lanes 4 and 9), or 2.7 μM (lanes 5 and 10) RuvBFH in the presence of Mg2+ (10 mM) and ATP (2 mM). After the reaction (5 min at 37°C), the samples were deproteinized, electrophoresed through native 12% polyacrylamide gels, and analyzed by fluorometry. The position of the substrate, which is too large to enter the gel, as well as the positions of the oligonucleotide reaction products, is indicated at the right-hand side of the gels by schematic illustrations. In these illustrations, the position of the 6-FAM label is indicated by a black circle. The asterisk points to DNA products in the gels that represent incorrectly annealed duplexes of unlabeled oligonucleotide 1, which is preferentially produced in the helicase reactions with substrate I, and labeled oligonucleotide 2 (see text). (C) The activity of RuvBMge on substrate I. Substrate I, 6-FAM labeled at the 5′ end of oligonucleotide 1, was incubated with either 0 μM (−; lane 1), 0.9 μM (lane 2), or 2.7 μM (lane 3) of RuvBMge. The other reaction parameters were similar to those described for panel A. (D) RuvAMpn does not influence the DNA helicase activity of RuvBFH. Substrate I, 6-FAM labeled at the 5′ end of either oligonucleotide 1 (lanes 1 to 5) or oligonucleotide 2 (lanes 6 to 10), was incubated without protein (lanes 1 and 6) or with 2.7 μM RuvBFH in the presence of either 0 nM (lanes 2 and 7), 19 nM (lanes 3 and 8), 56 nM (lanes 4 and 9), or 167 nM (lanes 5 and 10) RuvAMpn. The other reaction parameters were similar to those described for panel A. The asterisk points to DNA products in the gels that represent incorrectly annealed duplexes of unlabeled oligonucleotide 1 and labeled oligonucleotide 2. (E) RuvAMpn-independent helicase activity of RuvBFH and RuvAMge-dependent helicase activity of RuvBMge. The activities of RuvBFH (lanes 2 to 5) and RuvBMge (lanes 7 to 10) were tested on substrate IV in the presence of various concentrations of either RuvAMpn or RuvAMge (as indicated above the lanes) as follows: lanes 2 and 7, 0 nM; lanes 3 and 8, 56 nM; lanes 4 and 9, 167 nM; and lanes 5 and 10, 0.5 μM. The samples loaded in lanes 1 and 6 were incubated in the absence of any protein (−). The other reaction parameters were similar to those described in panel A.