Fig. 2.
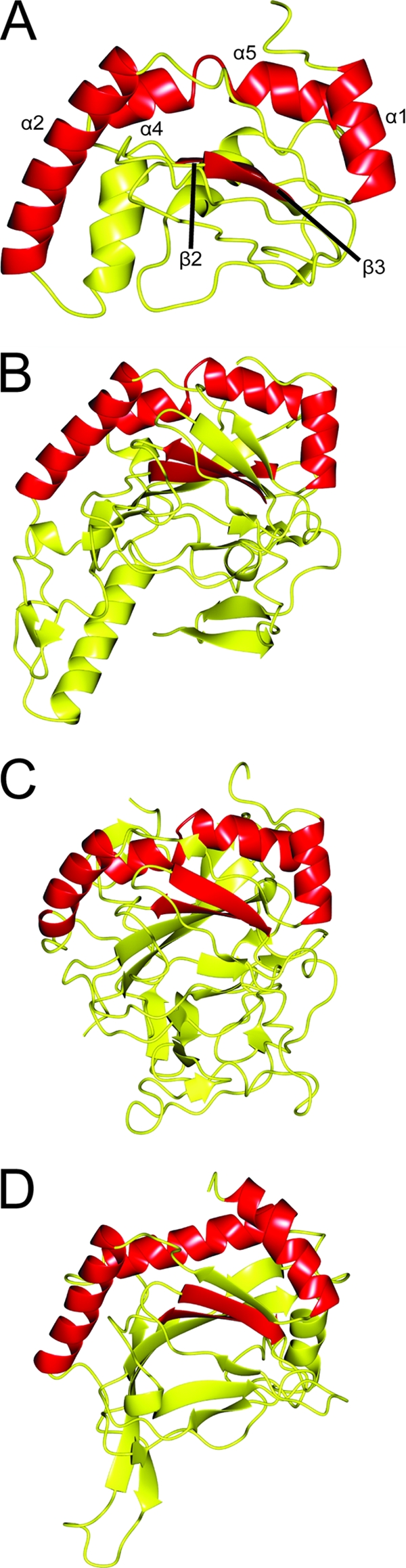
Ribbon depiction of the I-TASSER model for CT296 aligned with three proteins exhibiting the highest structural homology. Regions of structure that are conserved between all four structures are highlighted in red. (A) I-TASSER model of CT296. Four α-helices are highlighted (with residue numbers in parentheses): α1 (L7 to H16), α2 (V27 to F40), α3 (G65 to V73), and α4 (L75 to T83). Two β-strands are highlighted: β3 (D108 to C114) and β4 (W122 to F126). (B) Crystal structure of H. sapiens PHYD1 (PDB no. 2OPW). Four α-helices (similarly oriented to α1, α2, α3, and α4 from the I-TASSER model of CT296) and two β-strands (similar to β3 and β4 of CT296) are high-lighted. (C) Crystal structure of P. syringae SyrB2 (PDB no. 2FCU). Four α-helices (similarly oriented to α1, α2, α3, and α4 from the I-TASSER model of CT296) and two β-strands (similar to β3 and β4 of CT296) are highlighted. (D) Crystal structure of H. sapiens PHD2 (PDB no. 2HBT). Three α-helices (similarly oriented to α1, α2, and α3 + 4 from the I-TASSER model of CT296) and two β-strands (similar to β3 and β4 of CT296) are highlighted.
