Fig. 3.
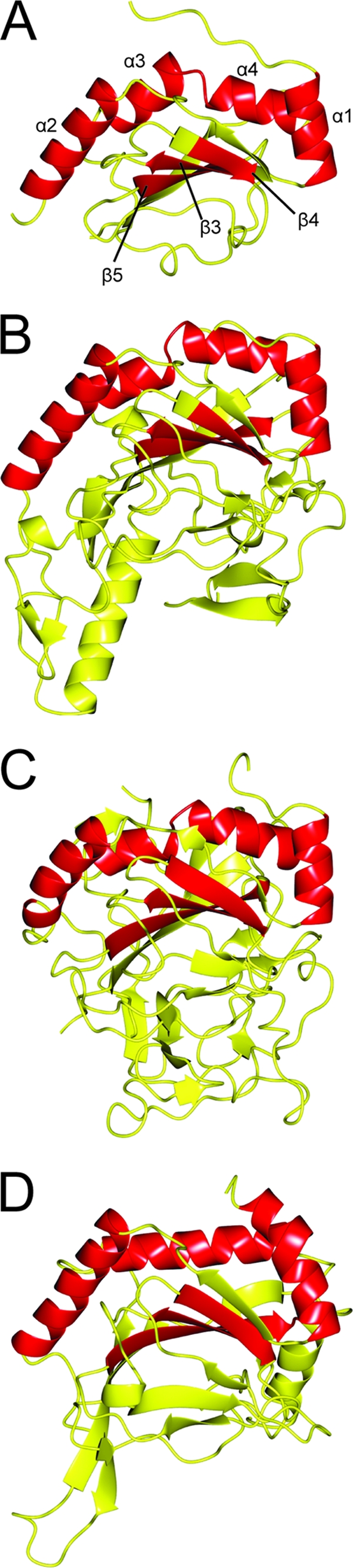
Ribbon depiction of experimentally determined structure of CT296 aligned with three proteins exhibiting the highest structural homology. Regions of structure that are conserved between all four structures are highlighted in red. (A) Crystal structure of CT296. Four α-helices are highlighted (with residue numbers in parentheses): α1 (L7 to H16), α2 (V27 to F40), α3 (G65 to V73), and α4 (L75 to T83). Three β-strands are highlighted (with residue numbers in parentheses): β3 (D108 to C114), β4 (W122 to F126), and β5 (A143 to S149). (B) Crystal structure of H. sapien PHYD1 (PDB no. 2OPW). Four α-helices (similarly oriented to α1, α2, α3, and α4 from the I-TASSER model of CT296) and three β-strands (similar to β3, β4, and β5 of CT296) are highlighted. (C) Crystal structure of P. syringae SyrB2 (PDB no. 2FCU). Four α-helices (similarly oriented to α1, α2, α3, and α4 from the I-TASSER model of CT296) and three β-strands (similar to β3, β4, and β5 of CT296) are highlighted. (D) Crystal structure of H. sapiens PHD2 (PDB no. 2HBT). Three α-helices (similarly oriented to α1, α2, and α3 + 4 from the I-TASSER model of CT296) and three β-strands (similar to β3, β4, and β5 of CT296) are highlighted.
