Abstract
Insulin exerts many of its metabolic actions via the canonical phosphatidylinositide 3 kinase (PI3K)/Akt pathway, leading to phosphorylation and 14-3-3 binding of key metabolic targets. We previously identified a GTPase-activating protein (GAP) for Rac1 called RhoGAP22 as an insulin-responsive 14-3-3 binding protein. Insulin increased 14-3-3 binding to RhoGAP22 fourfold, and this effect was PI3K dependent. We identified two insulin-responsive 14-3-3 binding sites (pSer16 and pSer395) within RhoGAP22, and mutagenesis studies revealed a complex interplay between the phosphorylation at these two sites. Mutating Ser16 to alanine blocked 14-3-3 binding to RhoGAP22 in vivo, and phosphorylation at Ser16 was mediated by the kinase Akt. Overexpression of a mutant RhoGAP22 that was unable to bind 14-3-3 reduced cell motility in NIH-3T3 fibroblasts, and this effect was dependent on a functional GAP domain. Mutation of the catalytic arginine of the GAP domain of RhoGAP22 potentiated growth factor-stimulated Rac1 GTP loading. We propose that insulin and possibly growth factors such as platelet-derived growth factor may play a novel role in regulating cell migration and motility via the Akt-dependent phosphorylation of RhoGAP22, leading to modulation of Rac1 activity.
INTRODUCTION
The serine/threonine kinase Akt acts as a central regulator of cellular metabolism downstream of insulin signaling. Phosphorylation of substrate proteins by Akt leads to changes in their enzymatic activity, localization, or protein-protein interactions and subsequently leads to alterations in physiology. The phosphorylation of many Akt substrates creates a binding motif for 14-3-3 protein family members, and for many substrates, 14-3-3 binding is responsible for translating Akt phosphorylation events into physiological outcomes (21). We recently reported the discovery of novel Akt substrates via the combination of 14-3-3 affinity chromatography and quantitative mass spectrometry (17). One of the proteins we identified was RhoGAP22, a product of the ARHGAP22 gene. RhoGAP22 is a GTPase-activating protein (GAP) for Rac1 and an inhibitor of VEZF1-dependent transcriptional activity (3). In a large-scale siRNA screen, knockdown of RhoGAP22 caused a defect in the transition from the ameboid to mesenchymal mode of cell movement, suggesting that ARHGAP22 has an important role in controlling cell motility (27). Binding of 14-3-3 regulates the GAP activity and function of other GAPs, including the RabGAP AS160 (25) and the RhoGAP DLC-1 (28). However, the involvement of 14-3-3 in regulating the activity of a Rac1 GAP has not previously been reported.
Akt plays a key role in cell migration through the activation of Rac1 (15). The generation of PI(3,4,5)P3 by phosphatidylinositol 3 kinase (PI3K) at the leading edge of a migrating cell is necessary for directed migration (16) and for lamellipodium formation in response to an attractant during chemotaxis (4). The presence of PI(3,4,5)P3 at the leading edge recruits and activates guanine nucleotide exchange factors (GEFs), such as Vav (1, 14), that promote the GTP loading and activation of Rac and CDC42. Akt binds PI3,4,5)P3 and may localize to the leading edge of the cell to regulate the activity of substrates that coordinate changes in cell motility, such as the actin-binding protein girdin (11) and PAKa (10). Indeed, the Rac effector protein Pak is also capable of targeting Akt to the plasma membrane, where it can be activated by PDK-1, and this pathway is important for cell migration in endothelial cells (23). Specific activation of Akt1 has also been found to decrease mammary epithelial cell migration, suggesting that Akt alone has substrates directly involved in controlling cell motility (30). Here, we report that RhoGAP22 is an Akt substrate that binds 14-3-3 in response to insulin. We have mapped the sites of Akt phosphorylation and 14-3-3 binding and determined that this plays a key role in regulating the GAP activity of RhoGAP22 and in regulated cell migration in NIH-3T3 cells.
MATERIALS AND METHODS
Materials.
Dulbecco's modified Eagle medium (DMEM), minimal essential medium (alpha) (αMEM), and F-12 nutrient mixture (Ham) were obtained from Invitrogen (Carlsbad, CA), Myoclone Plus fetal calf serum (FCS) was from Trace Scientific (Melbourne, Australia), and antibiotics were from Gibco BRL (Paisley, United Kingdom). Insulin was obtained from Calbiochem (San Diego, CA) and bovine serum albumin (BSA) from USB (Cleveland, OH). Bicinchoninic acid (BCA) reagent, Supersignal West Pico chemiluminescent substrate, Tris-carboxyethylphosphine (Bond-Breaker TCEP neutral pH solution), and protein G agarose beads were from Pierce (Rockford, IL). Lipofectamine LTX, Lipofectamine 2000 Plus reagent, and Sypro Ruby were from Invitrogen (Carlsbad, CA). Polyvinylidene difluoride (PVDF) membranes were from Millipore (Billerica, MA). Trypsin was from Promega (Madison, WI). Complete protease inhibitor cocktail tablets were from Roche (Indianapolis, IN). Magic C18 material was from Alltech (Deerfield, IL). Gateway cloning reagents, including pDONR221, pDEST vectors, and BP and LR clonases, were from Invitrogen. Unlabeled and stable-isotope-labeled arginine and lysine (>95% pure) were from Cambridge Isotope Laboratories (Andover, MA). All other materials were obtained from Sigma (St. Louis, MO). Pan-14-3-3 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Total Akt, Akt pS473, glutathione S-transferase (GST) antibodies, and recombinant Akt were purchased from Cell Signaling Technology (Danvers, MA). FLAG antibody (clone M2) was purchased from Sigma (St. Louis, MO). Alexa Fluor 680-conjugated secondary antibodies were obtained from Invitrogen. IrDye 800-conjugated secondary antibodies were obtained from Rockland Immunochemicals (Gilbertsville, PA). Horseradish peroxidase (HRP)-conjugated secondary antibodies and CNBr Sepharose were from GE Healthcare (Buckinghamshire, United Kingdom). [γ-32P]ATP was from MP Biomedicals (Solon, OH).
Plasmids and constructs.
Human RhoGAP22 cDNA was purchased from Open Biosystems (clone 8992160, corresponding to GenBank accession number AAI26445.1). This construct was amplified by using PCR to attach an N-terminal FLAG tag and 5′ and 3′ attB sites, allowing Gateway BP recombination into the pDONR221 vector (Invitrogen). FLAG-RhoGAP22 was inserted into a Gateway-converted pMIG internal ribosome entry sequence (IRES) green fluorescent protein (GFP) retroviral vector (7) using LR recombination. The S16A, R211A, S395A, and S476A mutants were created using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The S16A/S395A and S16A/S395A/R211A mutants were generated by restriction cloning using PstI/XmaI and XbaI/AgeI, respectively. RhoGAP22 fragments corresponding to amino acids 1 to 147 and 126 to 714 were amplified from the pDONR221 plasmid using PCR and inserted into pDEST53 (Invitrogen) to create N-terminal GFP fusions. Mouse RhoGAP22 cDNA was purchased from Open Biosystems (clone 4237331, corresponding to GenBank accession number AAH38272.1). This construct was amplified using PCR to create a fragment corresponding to amino acids 355 to 609 tagged with attB sites at the 5′ and 3′ ends. The PCR product was inserted into pDONR221 using BP recombination and cloned into pDEST15 and pDEST17 (Invitrogen) by LR recombination to generate N-terminal GST and 6-His fusions, respectively. Myc-tagged p68RacGAP was a gift from C. Patterson (3).
siRNA sequences.
RhoGAP22 specific and scramble control small interfering (siRNA) duplexes were from GenePharma (Shanghai, China). The sequences of the siRNAs used in this study were as follows: scramble control, 5′-UUCUCCGAACGUGUCACGUTT-3′; RhoGAP22 siRNA 1, 5′-GCGGAGGGAUCUUUGGGCATT-3′; RhoGAP22 siRNA 2, 5′-CAGUGCACCAUGAGCGGAATT-3′; RhoGAP22 siRNA 3, 5′-CUGGGAAGCUUGACUGCUGTT-3′.
Cell lines.
NIH 3T3 fibroblasts and L6 myoblasts were purchased from the American Type Culture Collection (ATCC, Rockville, MD). Chinese hamster ovary (CHO) cells stably expressing the insulin receptor and IRS-1 (CHO IR/IRS-1) were a gift from Morris White. Platinum E (Plat-E) cells were a gift from T. Kitamura (20). Double Akt1/2 knockout MEFs and wild-type controls were a gift from Morris Birnbaum (6).
Transient transfection of CHO IR/IRS-1 cells and L6 myoblasts using Lipofectamine LTX or Lipofectamine 2000.
Various FLAG-RhoGAP22 constructs were expressed in CHO IR/IRS-1 cells using Lipofectamine LTX as per the manufacturer's protocol. RhoGAP22-specific and control siRNAs were transfected into L6 myoblasts using Lipofectamine 2000 as per the manufacturer's protocol.
Retroviral infection of NIH 3T3 fibroblasts and generation of stable cell lines.
NIH 3T3 fibroblasts cultured in 10-cm dishes were incubated with pMIG retroviral supernatant in the presence of 4 mg/ml Polybrene. At 24 h postinfection, cells were sorted for GFP expression using a FACS Vantage SETM cell sorter (BD Biosciences). Cells expressing a moderate amount of GFP (median, 20%) were collected and used to establish a polyclonal cell line.
Production of RhoGAP22 antibody.
RhoGAP22-specific antisera were produced by IMVS Veterinary Services Division (Adelaide, SA, Australia). Briefly, mouse RhoGAP22 was amplified by PCR to produce a fragment corresponding to amino acids 355 to 609 and cloned into pDEST53 as described above to produce an N-terminally tagged GST fusion protein. Rabbits were immunized with 1 mg of antigen emulsified in Freund's complete adjuvant given subcutaneously at day 0. Subsequent immunizations consisted of 1 mg antigen emulsified in Freund's incomplete adjuvant given subcutaneously every 3 weeks for a total of 4 immunizations. Rabbits were exsanguinated at week 10.5, and serum was collected.
Production of the pSer22 and pSer397 RhoGAP22 antibody.
Antibodies were raised against peptides corresponding to amino acids 16 to 29 (Ac–YFTRSK[pS]LVMGEQSC–NH2 and Ac–CYFTRSK[pS]LVMGEQS–NH2) for Ser22 or 389 to 404 (Ac–LPTHRTS[pS]LDGPAAC–NH2 and Ac–CLPTHRTS[pS]LDGPAAA–NH2) for Ser397. of mouse RhoGAP22 and were produced by 21st Century Biochemicals (Marlboro, MA). Rabbits were inoculated with RhoGAP22 pSer22 or pSer397 peptides conjugated to keyhole limpet hemocyanin carrier protein five times over a 72-day period. Subsequently, the phospho-specific antibody was affinity purified by removal of antibodies that bind the nonphosphorylated peptide and then affinity purified using the phosphorylated peptide.
phospho-specific antibodies were produced in rabbits against the indicated peptides which correspond to the amino acid sequence of mouse RhoGAP22.
14-3-3 pulldown from L6 myotubes and immunoprecipitation using FLAG antibody.
14-3-3 pulldowns from 6-day L6 myotubes were performed as described previously (17). For anti-FLAG immunoprecipitation, cells expressing FLAG-RhoGAP22 constructs were serum starved for 2 h and then stimulated with 100 nM insulin for 20 min. Cells were washed three times on ice with ice-cold Tris-buffered saline (TBS). All liquid was removed, and cells were lysed in 1 ml of immunoprecipitation (IP) buffer (1% [vol/vol] Igepal CA-630, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10% [vol/vol] glycerol, and complete protease and phosphatase inhibitors) per dish. Lysates were passed 10 times through a 22-gauge needle and incubated on ice for 20 min, followed by centrifugation for 30 min at 18,000 × g at 4°C to pellet insoluble material. The supernatant was transferred to a new tube. Protein concentration was determined using a BCA assay as per the manufacturer's instructions. Equal amounts of protein from each sample were then incubated with 20 μl protein G-Sepharose that had been washed once in IP buffer and 1 μl of FLAG antibody. Samples were incubated overnight at 4°C on a rotating wheel. Beads were washed three times in cold IP buffer by resuspension and centrifugation at 2,000 g for 2 min at 4°C. Beads were washed once in ice-cold TBS, and all liquid was removed by aspiration with a microloader tip. FLAG-tagged proteins were eluted by addition of 50 μl TBS containing 200 μg/ml 3× FLAG peptide. Samples were incubated on ice for 1 h, with gentle agitation every 20 min. Following incubation, samples were centrifuged at 2,000 × g for 2 min at 4°C, and 40 μl of eluate was removed. Samples were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by addition of 4× SDS-PAGE buffer and TCEP to a final concentration of 50 mM.
Western blotting and SDS-PAGE.
SDS-PAGE analysis was performed on 10% or 7.5% resolving gels with the addition of 50 mM TCEP in the sample buffer. Equal amounts of protein were loaded for each sample in a single experiment, typically 10 μg per lane. For mass spectrometric identification, Sypro Ruby staining was performed as per the manufacturer's instructions (Invitrogen). For Western blotting, proteins were electrophoretically transferred to PVDF membranes, and the membrane was blocked with 5% nonfat milk in 0.1% (vol/vol) Tween 20 in TBS (TBST) and incubated with primary antibody in 5% BSA in TBST overnight at 4°C. After incubation, membranes were washed three times in TBST and incubated with HRP-labeled or Alexa fluor 680/IrDye 800-labeled secondary antibodies in 5% nonfat milk in TBST for HRP-conjugated secondary antibodies or TBST with 0.01% SDS (wt/vol) for fluorescent secondary antibodies. Proteins were visualized using Supersignal West Pico chemiluminescent substrate and imaged with X-ray film (Fuji) for HRP-labeled secondary antibodies or a Licor Odyssey imager for Alexa fluor 680/IrDye 800-labeled secondary antibodies.
In-gel tryptic digest for peptide identification by LC-MS/MS.
FLAG-RhoGAP22 was transiently expressed in CHO IR/IRS-1 cells as described above, immunoprecipitated using FLAG antibody from either basal or insulin-stimulated cells (100 nM, 30 min), subjected to 10% SDS-PAGE, and stained with Sypro Ruby. Protein bands of interest were excised and destained in 1 ml of 50% acetonitrile, 250 mM NH4HCO3 at room temperature (RT) for 45 min with shaking. The gel slice was dehydrated by incubation in 1 ml of 100% acetonitrile for 10 min at RT. All solution was carefully removed using a microloader tip prior to the addition of modified trypsin (12.5 ng/μl) in 100 mM NH4HCO3 and incubation overnight at 37°C. The following day, peptides were extracted by the addition of 0.1 ml of 5% formic acid and incubation at 37°C for 1 h. Peptides were further extracted by the addition of 0.1 ml of 100% acetonitrile and incubation at 37°C for 1 h. The gel slice was completely dehydrated by the addition of 0.5 ml of 100% acetonitrile and incubation at 37°C for 10 min. The entire supernatant was then removed, transferred to a new tube, and vacuum dried. Peptides were redissolved in 20 μl of 5% formic acid for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Peptide identification by LC-MS/MS was performed as described previously (17).
In vitro phosphorylation using recombinant Akt.
Various FLAG-GFP-tagged RhoGAP22 constructs were expressed in HEK cells using Lipofectamine 2000 as per the manufacturer's instructions. Cells were cultured for 48 h and then treated with 100 nM wortmannin for 30 min. Cells were lysed on ice, and proteins were immunoprecipitated using FLAG antibody as described above but with RIPA buffer (0.1% [vol/vol] SDS, 0.5% [wt/vol] sodium deoxycholate, 1% [vol/vol] Igepal CA-630, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, EDTA-free complete protease inhibitors and phosphatase inhibitors) and natively eluted using 3× FLAG peptide. Eluate (9 μl) was transferred to a new tube and combined with 5 μl of 3 × assay buffer (75 mM Tris-HCl [pH 7.4], 6 mM dithiothreitol, 30 mM MgCl2), 5 μl of ATP mix (14.8 kBq/μl [γ-32P]ATP in 800 μM ATP dissolved in 1× assay buffer) and 1 μl diluted Akt kinase (200 ng/μl in 1× assay buffer). Reaction mixtures were incubated at room temperature for 10, 30, or 60 min and stopped by addition of 4× SDS-PAGE buffer to a 1× concentration and TCEP to 50 mM and heated at 65°C for 5 min. Entire sample was subjected to 10% SDS-PAGE and then fixed and stained with Coomassie brilliant blue dye for 30 min. The gel was destained, equilibrated using MilliQ water, and dried in a vacuum gel drier onto filter paper. The dried gel was exposed to a phospho-imager plate overnight, and the plate was imaged using a Fujifilm FLA-500 imaging system (Fujifilm, Tokyo, Japan).
Cell motility assays.
NIH 3T3 fibroblasts stably overexpressing various RhoGAP22 mutants by pMIG-GFP infection were mixed in equal amounts with NIH 3T3 fibroblasts infected with a pMIG-mCherry construct and seeded into fibronectin (10 μg/ml in PBS for 60 min)-coated 24-well plates at a final concentration of 5,000 cells/ml. Cells were cultured overnight at 37°C in 10% CO2 using a heated CO2 incubator attachment to a Zeiss Observer inverted microscope (Carl Zeiss, Oberkochen, Germany). Epifluorescent images in both the green and red channels were taken every 10 min for about 14 h using a 10× objective. Image stacks were minimally processed in Image J (2) using a Gaussian blur and rolling ball algorithm to reduce background. Processed images were analyzed using Imaris software (Bit Plane AG, Zurich, Switzerland). Cells were assigned spots in each frame of the image stack, and spots were then tracked over time using the Imaris particle tracking module. To qualify for analysis, particles were required to have a minimum size of 9.5 μm and a quality of >10 (as defined by the Imaris software) and had to be present for at least 30 consecutive frames (5 h). The total displacement of each cell in the stack over time was calculated and expressed as the mean ratio of the displacement of RhoGAP22-expressing cells (green) over the displacement of pMIG mCherry control cells (red). Significance was determined by one-way repeated-measures analysis of variance (ANOVA), followed by Tukey's multiple comparison test.
GST-PAK pulldown assay.
Rac-GTP loading was determined using a GST pulldown approach utilizing the CDC42/Rac interactive binding (CRIB) domain of p21-activated kinase (PAK) produced as a GST fusion protein (PAK PBD). Subconfluent NIH 3T3 fibroblasts stably overexpressing various RhoGAP22 mutants by pMIG-GFP infection were serum starved for 2 h and stimulated with 20 ng/ml platelet-derived growth factor (PDGF) for 5 min. Cells were washed three times on ice with ice-cold HEPES-buffered saline (HBS). All liquid was removed, and cells were lysed in assay buffer (25 mM HEPES [pH 7.4], 150 mM NaCl, 1% [vol/vol] Igepal CA-630, 0.25% [wt/vol] Na deoxycholate, 10% [vol/vol] glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 10 mg/ml leupeptin, and 10 mg/ml aprotinin). Lysates were passed 10 times through a 22-gauge needle and incubated on ice for 15 min, followed by centrifugation for 15 min at 18,000 × g at 4°C to pellet insoluble material. The supernatant was transferred to a new tube. Protein concentration was determined using a BCA assay as per the manufacturer's instructions. Equal amounts of protein from each sample were then incubated with freshly prepared GST-PAK PBD beads that had been washed three times with assay buffer. Samples were incubated for 20 min at 4°C on a rotating wheel. Beads were washed three times in cold assay buffer by resuspension and centrifugation at 2,000 × g for 2 min at 4°C. All liquid was removed by aspiration with a microloader tip and eluted in 1× SDS-PAGE sample buffer containing 50 mM TCEP.
Immunofluorescence.
L6 myoblasts were cultured on Matrigel-coated glass coverslips and transfected with either 20 pmol of scramble control siRNA or 6.66 pmol of each RhoGAP22-directed siRNA in combination as described above. Cells were cultured for 48 h, serum starved for 2 h, and treated with 100 nM insulin as indicated in Fig. 6. Cells were washed 3× on ice with ice-cold PBS. All liquid was removed, and cells were fixed in 3% paraformaldehyde in PBS for 30 min at RT before quenching with 50 mM glycine in PBS for 20 min. Cells were blocked and permeabilized in blocking buffer (2% BSA and 0.1% saponin in PBS) (BB) for 30 min with shaking. Permeabilized cells were incubated with Alexa 647-conjugated phalloidin in BB for 1 h at RT. Coverslips were washed eight times in PBS and then mounted in mounting medium. Optical sections were analyzed by confocal laser scanning microscopy using a Leica TCS SP system using a 60× lens. Cellular morphology was quantitated using Cell Profiler software (8).
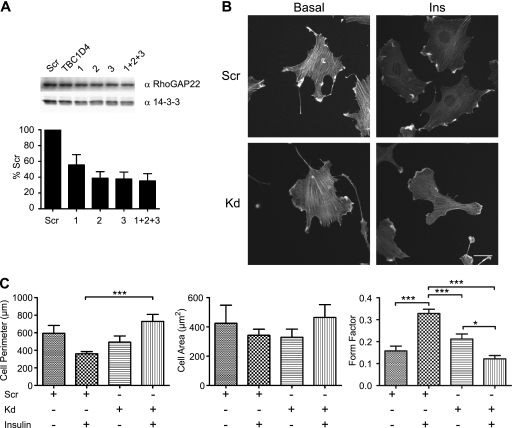
Fig. 6.
Knockdown of endogenous RhoGAP22 levels causes defects in insulin-regulated morphological changes. (A) Knockdown of RhoGAP22 in L6 myoblasts using siRNA. L6 myoblasts were transfected with 20 pmol of nontargeting control siRNA (Scr), 20 pmol of siRNA directed against an unrelated protein (TBC1D4), or either 20 pmol of RhoGAP22 siRNAs individually (1, 2, and 3) or 6.66 pmol of each RhoGAP22 siRNA pooled (1+2+3). Cells were cultured for 48 h prior to lysis and Western blotting with indicated antibodies. (B) RhoGAP22 knockdown caused defects in insulin-regulated morphological change. L6 myoblasts were transfected with either control siRNA (Scr) or pooled RhoGAP22 siRNAs (Kd). Cells were cultured for 48 h prior to serum starvation and treatment (Ins) or no treatment (Basal) with 100 nM insulin for 20 min. Cells were fixed and the actin cytoskeleton stained with Alexa 647 phalloidin. Representative images are shown. (C) Quantitation of defective morphological change in RhoGAP22 knockdown cells. Micrographs from panel B were analyzed using the Cell Profiler software package. At least 15 cells were analyzed in each condition. Error bars indicate standard errors of the means. Significance was determined by one-way ANOVA followed by Tukey's posttest. *, P < 0.05; ***, P < 0.001.
RESULTS
RhoGAP22 is an insulin-responsive 14-3-3 binding protein.
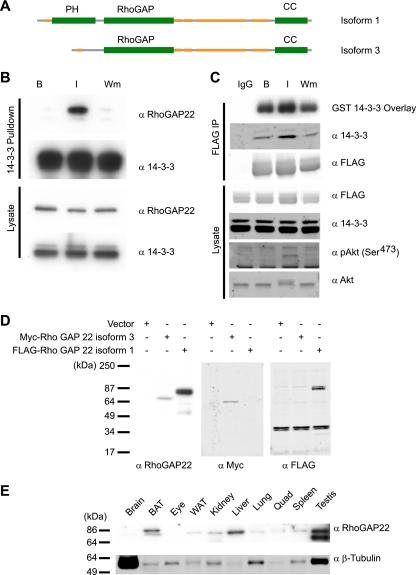
Previously we showed that the binding of RhoGAP22 to 14-3-3 was increased fourfold in response to insulin stimulation and that this interaction was dependent on PI3K signaling (17). RhoGAP22 has previously been reported as a GTPase-activating protein with specific activity for Rac1, a master regulator of the actin cytoskeleton and cell motility. RhoGAP22 contains three conserved protein domains, as predicted by sequence homology using the SMART tool (18): an N-terminal pleckstrin homology (PH) domain (residues 38 to 147); a Rho GAP domain, defining it as a member of the Rho GTPase-activating protein family (residues 140 to 376); and a C-terminal coiled-coil domain (residues 607 to 687) thought to be important for protein-protein interactions (Fig. 1A). Analysis of protein disorder by the GlobPlot algorithm (19) indicates that the N terminus and a large central region between the RhoGAP domain and the coiled-coil domain are largely disordered. To enable further analysis, we generated an antibody specific for RhoGAP22 recognizing the disordered region between the Rho GAP and coiled-coil domains. This region has the least homology to other Rho GAPs and is common to all RhoGAP22 isoforms. Hence, our prediction was that an antibody raised against this region would recognize all predicted RhoGAP22 isoforms. To test our antibody, we immunoblotted lysates from cells expressing vector, recombinant Myc-tagged human RhoGAP22 isoform 2, or FLAG-tagged mouse RhoGAP22 isoform 1 (Fig. 1D). The antibody recognized both RhoGAP22 isoforms, as shown by labeling of bands at 67 and 77 kDa, corresponding to the predicted molecular masses of each isoform. Immunoblotting of a range of mouse tissue lysates revealed RhoGAP22 to be broadly expressed, with the highest levels of expression being found in brown adipose tissue, liver, and testis (Fig. 1E). The predominant band identified had an average molecular mass of ∼77 kDa, which likely corresponds to RhoGAP22 isoform 1. Testes also displayed a prominent band at 67 kDa, consistent with abundant expression of isoform 3 in this tissue. Next, we wanted to confirm the insulin-responsive 14-3-3 interaction with RhoGAP22 in a cellular model of insulin action. To this end, an in vitro 14-3-3 pulldown was performed on lysates from serum-starved L6 myotubes incubated with or without insulin or insulin combined with wortmannin pretreatment. There was an increase in endogenous RhoGAP22 isoform 1 associated with 14-3-3 in response to insulin stimulation, and this interaction was inhibited with wortmannin (Fig. 1A). To confirm that this was a direct interaction, we expressed full-length recombinant FLAG-tagged RhoGAP22 in CHO IR/IRS-1 cells. Immunoprecipitation of this construct followed by far-Western blotting with recombinant GST–14-3-3 showed that insulin induced a PI3K-dependent increase in the direct binding of RhoGAP22 to 14-3-3 (Fig. 1C). Moreover, we also observed an insulin- and PI3K-dependent coprecipitation of endogenous 14-3-3 with RhoGAP22 in this experiment (Fig. 1C). Interestingly, there was significant binding of 14-3-3 to RhoGAP22 in the basal state, indicating the presence of a constitutive 14-3-3 binding site in RhoGAP22.
Fig. 1.
RhoGAP22 binds 14-3-3 in response to an insulin stimulus and production of a RhoGAP22 antibody. (A) Schematic representation of the domain organization of RhoGAP22 isoform 1. PH, pleckstrin homology domain; RhoGAP, Rho GTPase-activating domain; CC, coiled-coil domain. Yellow indicates regions of low complexity. (B) Endogenous RhoGAP22 binds 14-3-3 in an insulin- and PI3K-dependent manner. L6 myotubes were serum starved (B) and treated with 100 nM insulin for 30 min (I) or insulin and 100 nM wortmannin (Wm). Cell lysates were subjected to 14-3-3 pulldown and immunoblotted with indicated antibodies. (C) Overexpressed RhoGAP22 binds 14-3-3 similarly to the endogenous protein. FLAG-RhoGAP22 was expressed in CHO IR/IRS-1 cells. Cells were serum starved and treated as basal (IgG and B) or treated with 100 nm insulin for 30 min (I) or with 100 nM wortmannin for 30 min and then 100 nM insulin for 30 min (Wm). Cells lysates were immunoprecipitated with either normal IgG as a control (IgG) or anti-FLAG antibody. (D) The RhoGAP22 antibody specifically recognizes various isoforms of RhoGAP22. CHO IR/IRS-1 cells were transfected with the indicated RhoGAP22 constructs or empty vector. Cell lysates were blotted with the indicated antibodies. (E) Various mouse tissue lysates (10 μg per lane) were blotted with the indicated antibodies. BAT, brown adipose tissue; WAT, white adipose tissue; Quad, quadriceps.
pS16 and pS395 mediate insulin-stimulated 14-3-3 binding to RhoGAP22.
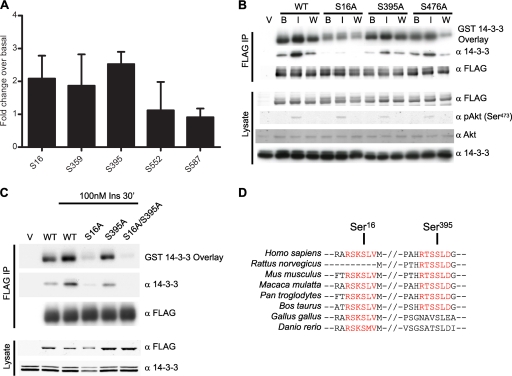
We next sought to determine the 14-3-3 binding site(s) in RhoGAP22. Scansite predicted three medium-stringency mode 1 14-3-3 sites at Ser16, Thr424, and Thr615 (all within the top 0.75% of all mode 1 14-3-3 sites in the Swiss-Prot vertebrate database) (22). The ELM tool predicted six additional mode 2 and 3 14-3-3 binding sites at Thr160, Ser395, Ser437, Ser439, Ser476, and Thr491; however, ELM does not provide expectation values (24). There are three medium-stringency Akt sites at Ser16, Thr189, and Ser395, as predicted by Scansite (all within the top 1% of all Akt sites in the Swiss-Prot vertebrate database). Thus, the only predicted 14-3-3 sites also predicted to be Akt sites were Ser16 and Ser395. To determine the possible role of these sites in insulin-regulated 14-3-3 binding, we employed semiquantitative mass spectrometry. This approach overcame the need to mutate each of the eight potential 14-3-3-binding sites individually. FLAG-RhoGAP22 was immunoprecipitated from CHO IR/IRS-1 cells that had been serum starved and treated with insulin or left untreated. The immunoprecipitate was subjected to SDS-PAGE, and bands corresponding to FLAG-RhoGAP22 were subjected to in-gel tryptic digestion and LC-MS/MS. By comparing the abundance of each phosphopeptide with that of its cognate nonphosphopeptide for each condition, a relative semiquantitative measure of the change (fold) with insulin was obtained (7). The supplemental material presents the annotated results of this analysis (see Table S1-1 in the supplemental material), as well as mass spectra for selected phosphopeptides of putative 14-3-3 binding sites (see Fig. S1 and S2 in the supplemental material). Figure 2A shows a summary of these data, displayed as the mean relative fold change in phosphorylation at each site corrected by each cognate nonphosphopeptide. Of the five phosphopeptides identified, only Ser16, Ser359, and Ser395 exhibited an increase in phosphorylation with insulin. However, only Ser16 and Ser395 are predicted to form part of a potential 14-3-3 binding site. Phosphopeptides comprising a third potential 14-3-3 binding site at Ser476 were also identified but were not included in the analysis due to the presence of a missed cleavage at an N-terminal arginine close to the phosphoserine. This gave rise to alternate peptide species, rendering it unsuitable for this kind of analysis. To confirm that Ser16 and Ser395 were the insulin-responsive 14-3-3 binding sites, we used site-directed mutagenesis to mutate each serine to alanine. Each mutant, as well as the wild-type protein, was immunoprecipitated from CHO IR/IRS-1 cells that had been serum starved and treated with or without insulin or insulin combined with wortmannin pretreatment. The ability of each protein to bind 14-3-3 in vivo and in vitro was determined by Western blotting for coimmunoprecipitated endogenous 14-3-3 as well as direct 14-3-3 binding via far-Western blotting with GST 14-3-3. Figure 2B shows that the S16A mutation greatly reduced insulin-responsive in vivo binding to 14-3-3. The S16A mutant was still competent to bind 14-3-3 in vitro, although this binding appeared to be insulin and wortmannin insensitive. The S395A mutant displayed reduced in vivo binding to 14-3-3, but not to the same extent as the S16A mutant. This mutant also bound 14-3-3 in vitro in an insulin- and wortmannin-dependent fashion. The S476A mutant had only slightly reduced in vivo binding to 14-3-3, and yet this mutation had a greater effect on 14-3-3 binding in vitro than the S395A mutation. All three sites contributed to 14-3-3 binding in vitro, as indicated by the reduction in binding compared to the wild-type protein. These results suggested that more than one phosphorylation site is responsible for the observed insulin-responsive 14-3-3 binding in vivo and that the in vitro far-Western analysis for direct 14-3-3 interaction may not give a true reflection of in vivo 14-3-3 binding. To test the multisite binding hypothesis, a FLAG-tagged S16A/S395A mutant was generated and immunoprecipitated as before. This mutant was unable to bind 14-3-3 in vivo (Fig. 2C) and bound only trace amounts of 14-3-3 in vitro. This suggests that both Ser16 and Ser395 are required for insulin-responsive 14-3-3 binding, although the stoichiometry of the interaction is unclear. The residual amount of 14-3-3 that bound in vitro was probably bound to the Ser476 site, although this site did not contribute to binding in vivo. Sequence alignment using the BLAST algorithm indicated that both the Ser16 and Ser395 binding sites are well conserved across species (Fig. 2D).
Fig. 2.
RhoGAP22 binds 14-3-3 at both Ser16 and Ser395. (A) The phosphorylation of FLAG-RhoGAP22 under basal and insulin-stimulated conditions was analyzed by semiquantitative mass spectrometry. Results are shown as the mean increase in phosphopeptide abundance, expressed as the fold change over the basal level. Values are means from three experiments; error bars indicate standard deviations. Residues on RhoGAP22 were mutated either singly (B) or in combination (C) to determine the insulin-responsive 14-3-3 binding site. FLAG-tagged proteins were overexpressed in CHO IR/IRS-1 cells. Cells were serum starved and treated with no additions (B), with 100 nM insulin for 30 min (I), or with 100 nM wortmannin for 30 min and then 100 nM insulin for 30 min (W). Cell lysates were subjected to FLAG immunoprecipitation and immunoblotted with the indicated antibodies or used in a far-Western blot with GST–14-3-3 as a probe. (D) Conservation of 14-3-3 binding sites of RhoGAP22 across various species. Red text indicates a putative 14-3-3 binding site. Sequence numbering refers to human RhoGAP22 isoform 1.
Phosphorylation at S16 and S395 is Akt dependent.
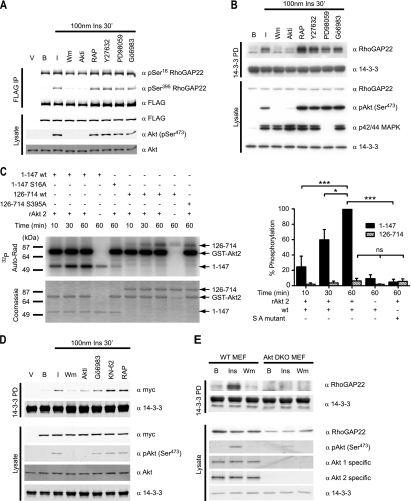
To identify the kinase(s) that regulates phosphorylation and 14-3-3 binding of RhoGAP22, we employed several kinase inhibitors in conjunction with immunoblotting using phospho-specific RhoGAP22 antibodies. These antibodies were directed against pSer16 and pSer395 and were found to be specific in recognizing their cognate phosphorylation sites by immunoblotting of immunoprecipitates and lysates from CHO IR/IRS-1 cells expressing wild-type and mutant RhoGAP22 (see Fig. S3 in the supplemental material) as well as in phosphopeptide competition assays (see Fig. S4 in the supplemental material). Kinases selected for inhibition were involved in the insulin signaling pathway (wortmannin, Akti, rapamycin, PD98059, and Gö6983) or in the control of Rho kinase (ROCK) signaling (Y27632). FLAG-RhoGAP22 was immunoprecipitated from CHO IR/IRS-1 cells that had been serum starved and incubated with or without insulin or with insulin and pretreatment with the various kinase inhibitors. The immunoprecipitates were immunoblotted using the phospho-RhoGAP22 antibodies as well as anti-FLAG antibodies. Phosphorylation at Ser16 was reduced when cells were pretreated with wortmannin or with Akti (a highly selective Akt inhibitor), although to a lesser extent than that observed with wortmannin. Pretreatment with wortmannin and Akti also reduced phosphorylation at Ser395. Neither Ser16 phosphorylation nor Ser395 phosphorylation was inhibited by pretreatment with an mTORC1 inhibitor (rapamycin), an inhibitor of the ROCK family of kinases (Y27632), an inhibitor of MEK1 (PD98059), or an inhibitor of various members of the PKC family (Gö6983) (Fig. 3A; also, see Fig. S5 in the supplemental material). Similarly, endogenous RhoGAP22 was unable to bind 14-3-3 in a pulldown assay from L6 myotubes that had been treated with either wortmannin or Akti but not other inhibitors (Fig. 3B). While we previously showed that the kinase responsible for regulating the binding of 14-3-3 to RhoGAP22 was downstream of PI3K via the use of wortmannin, this new result places the kinase at the level of Akt or a kinase downstream of Akt. To demonstrate that RhoGAP22 is a bona fide substrate of Akt, we used recombinant GST-Akt to phosphorylate FLAG-GFP-tagged fragments of RhoGAP22 in vitro. The full-length protein was not analyzed, as it is the same size as the GST-Akt used in the assay and cannot be resolved using SDS-PAGE. These data show that Akt phosphorylates the N-terminal RhoGAP22 truncation mutant (consisting of residues 1 to 147) in vitro, in a time-dependent manner (Fig. 3C; also, see Fig. S6 in the supplemental material). Phosphorylation of the 1–147 N-terminal fragment was substantially, although not completely, reduced when Akt was omitted from the assay. This residual phosphorylation may reflect the presence of a small amount of Akt coimmunoprecipitating with the 1-147 N-terminal fragment. Phosphorylation was totally absent in the S16A mutant indicating that this was the only site of Akt phosphorylation in this polypeptide. The FLAG-GFP-126-714 construct also exhibited a time-dependent increase in phosphorylation (Fig. 3C). However, phosphorylation of this construct was not reduced by S395A mutation indicating that phosphorylation likely also occurs at an alternate site. As with the 1-147 N-terminal fragment, residual phosphorylation was again seen in the 126-714 fragment when Akt was omitted from the reaction. Again, this is probably due to the presence of a small amount of coimmunoprecipitating Akt or another kinase. Statistically, there was no significant difference in the degree of phosphorylation of the 126–714 construct when Akt was omitted from the reaction or in the context of the S395A mutation. Thus, we concluded that Akt does not phosphorylate Ser395 in vitro. This was surprising, as we also used Myc-tagged RhoGAP22 isoform 2 in a kinase inhibitor screen similar to that shown in Fig. 3B. This construct lacks the N-terminal portion of the protein, including the Ser16 14-3-3 binding site. Using this construct, it was apparent that only wortmannin and Akti inhibited insulin-stimulated 14-3-3 binding (Fig. 3D). This result indicates that inhibition of Akt prevents phosphorylation at Ser395, as well as 14-3-3 binding to this site. It would seem that this site is not phosphorylated by Akt, although active Akt is required for its phosphorylation. These data illustrate that RhoGAP22 can be phosphorylated by Akt at one of the 14-3-3 binding sites and that inhibition of Akt prevents phosphorylation at both sites. To confirm that Akt was indeed required for phosphorylation of RhoGAP22, the 14-3-3 pulldown experiments were recapitulated in Akt 1/2 knockout mouse embryonic fibroblasts (6). These cells were serum starved and then treated with insulin, left untreated, or treated with insulin combined with wortmannin pretreatment, and cell lysates were subjected to 14-3-3 pulldown. Insulin-stimulated 14-3-3 binding to RhoGAP22 in the absence of Akt 1 and 2 was almost totally absent (Fig. 3E). There was still a very small amount of binding, probably caused by Akt 3 kinase activity, the presence of which could be inferred from the trace amount of Akt phosphorylated at Ser473 in these cells. This result further confirmed the requirement for Akt in the insulin-stimulated phosphorylation and 14-3-3 binding to RhoGAP22.
Fig. 3.
Akt phosphorylates RhoGAP22 at the 14-3-3 binding site and is required for insulin-stimulated 14-3-3 binding. (A) Insulin-stimulated phosphorylation of Ser16 and Ser395 is inhibited by wortmannin and Akti. CHO IR/IRS-1 cells expressing FLAG-RhoGAP22 were serum-starved and either left untreated (B), treated with 100 nM insulin for 30 min (I), or treated with 100 nM insulin for 30 min after one of the following: 100 nM wortmannin for 30 min (Wm), 5 μM Akti for 30 min (Akti), 20 nM rapamycin for 30 min (RAP), 10 μM Y27632 for 30 min (Y27632), 50 μM PD98059 for 30 min (PD98059), or 100 nm Gö6983 for 30 min (Gö6983). Treatment was followed by lysis and Western blotting with the indicated antibodies. (B) Insulin-stimulated 14-3-3 binding to endogenous RhoGAP22 is inhibited by wortmannin and Akti. L6 myotubes were serum starved and treated (I) or not treated (B) with 100 nM insulin for 30 min or with 100 nM insulin for 30 min after treatment with the indicated kinase inhibitors. (C) Recombinant Akt2 can phosphorylate Ser16 of RhoGAP22 in vitro. FLAG-GFP-tagged RhoGAP22 constructs were expressed in HEK cells by transfection with Lipofectamine 2000. Cells were treated with 100 nM wortmannin for 30 min, lysed, and subjected to FLAG immunoprecipitation. Eluted proteins were diluted in reaction buffer and incubated with 74 kBq [γ-32P]ATP with or without 200 ng of recombinant Akt2 as indicated and incubated for various times. Reactions were run on a 10% SDS-PAGE gel, and products were dried on filter paper. Incorporation of 32P was measured using a phospho-imager (Auto-Rad). The gel was stained with Coomassie blue to show total protein loading (Coomassie). Quantitation of these data are shown on the right as percentages of the phosphorylation of the 1-147 N-terminal fragment at the 60-min data point, normalized for protein loading. Significance was determined by one-way ANOVA followed by Tukey's posttest. *, P < 0.05; ***, P < 0.001; ns, not significant. (D) Wortmannin and Akti inhibit insulin-stimulated 14-3-3 binding to a RhoGAP22 construct that lacks the Ser16 site. CHO IR/IRS-1 cells were transfected with Myc-tagged p68RacGAP, serum starved, and treated (I) or not treated (B) with 100 nM for 30 min insulin or with 100 nM insulin for 30 min after treatment with the indicated kinase inhibitors. (E) Akt is required for insulin-stimulated 14-3-3 binding to RhoGAP22. Embryonic fibroblasts from either wild-type mice (WT MEF) or Akt 1/2 knockout mice (Akt DKO MEF) were serum starved and treated (Ins) or not treated (B) with 100 nm insulin for 30 min or with 100 nM wortmannin for 30 min and then 100 nM insulin (Wm). Cells were lysed, and lysates were used in a 14-3-3 pulldown before being immunoblotted with the indicated antibodies.
Phosphorylation of RhoGAP22 at S16 and S395 regulates cell motility.
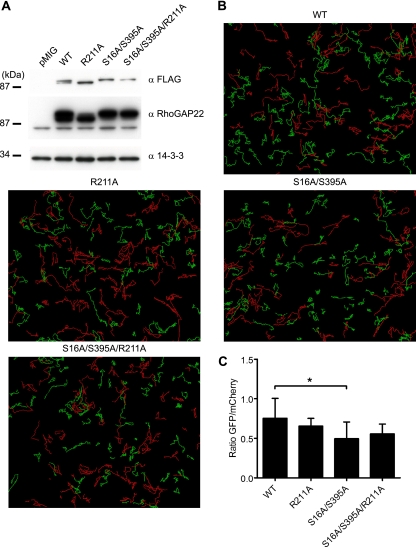
Having identified RhoGAP22 as an insulin-responsive 14-3-3 binding protein, we were interested in identifying the functional outcome of the insulin-stimulated 14-3-3 interaction with RhoGAP22. It has previously been shown using RNA interference that RhoGAP22 plays an important role in regulating the switch between the mesenchymal and amoeboid modes of cell migration, presumably by regulating the activity of Rac1 (27). To investigate the importance of the Rho GAP22 and 14-3-3 interaction in the control of Rac1 activity, we utilized a cell migration assay. NIH 3T3 fibroblasts stably expressing various RhoGAP22 constructs were generated by infection with pMIG retrovirus empty vector or pMIG retrovirus encoding FLAG-RhoGAP22 or FLAG-RhoGAP22 mutants. The pMIG retrovirus uses an internal ribosome entry sequence (IRES) to express GFP independently of the cDNA cloned into the multiple cloning site. We exploited this function to generate polyclonal cell lines using fluorescence-activated cell sorting (FACS). Infected NIH 3T3 fibroblasts were sorted, and the cells expressing the median level (±10%) of GFP (as measured by intensity of GFP signal) were collected and used to establish polyclonal cell lines. As GFP expression correlated with FLAG-RhoGAP22 expression, this provided an easy method for generating cell lines that were well matched for the level of FLAG-RhoGAP22 expression (Fig. 4A). Cell lines expressing empty pMIG vector (pMIG), wild-type RhoGAP22 (WT), RhoGAP22 where the critical catalytic residue in the GAP domain was mutated (R211A), a 14-3-3-binding mutant (S16A/S395A), and an S16A/S395A/R211A RhoGAP22 mutant were generated. The apparent difference in molecular weight of the mutant proteins was probably due to a posttranslational modification, as these constructs were completely sequenced and found to be correct. Mutation of the catalytic arginine did not affect insulin-stimulated 14-3-3 binding to RhoGAP22 (see Fig. S7 in the supplemental material). The NIH 3T3 cells were combined in equal numbers with NIH 3T3 cells stably expressing empty pMIG vector that also expressed mCherry under the control of the IRES, which would act as an internal control. These cells were plated onto a fibronectin-coated 24-well plate and imaged every 10 min for 14 h. Representative time-lapse videos for each cell line are available in Movie S1 in the supplemental material. Individual cells were tracked over the 14-h time course of the experiment. These tracks can be seen in Fig. 4B. The tracks for control cells expressing pMIG mCherry are represented by a red line; that of the cells expressing the FLAG-RhoGAP22 constructs are represented by a green line. There was no significant difference in the displacement of the control cells across the experiment (see Fig. S8 in the supplemental material). The abundance of shorter tracks for the S16A/S395A mutant indicated a defect in migration. However, there was no significant difference in total track length between cells expressing FLAG-RhoGAP22 constructs and control cells (data not shown). While the total track length is of interest, a cell may either migrate in a straight line or move in a more random fashion and still produce a track of the same length. It is therefore more interesting to study the directionality of its motion by examining its total displacement (12). The mean square displacement over 14 h was quantified, and results were expressed as the mean ratio of the mean displacement of the experimental cells lines to the mean displacement of the control cells. This showed that cells overexpressing a mutant FLAG-RhoGAP22 that was unable to bind 14-3-3 (S16A/S395A) had a significant reduction in displacement compared to cells expressing wild-type RhoGAP22 (Fig. 4C). In contrast, cells overexpressing mutant FLAG-RhoGAP22 that was unable to bind 14-3-3 and was also catalytically inactive (S16A/S395A/R211A) did not differ significantly from the wild type, indicating that the effect of loss of 14-3-3 binding was dependent on the GAP activity of RhoGAP22.
Fig. 4.
Overexpression of a mutant RhoGAP22 that cannot bind 14-3-3 results in decreased random cell motility. (A) NIH 3T3 fibroblasts were infected with pMIG retrovirus encoding empty vector (pMIG), FLAG-RhoGAP22 (WT), or mutants of FLAG-RhoGAP22. The pMIG retrovirus also independently expresses eGFP using an IRES. Cells were subjected to FACS for the median 20% of GFP-expressing cells of each line. Cells from each line were lysed, and lysates were immunoblotted with the indicated antibodies. (B) NIH 3T3 fibroblasts expressing various RhoGAP22 constructs (green) or vector only (red) were imaged every 10 min for 14 h. Tracks depict migration of individual cells. (C) Cell displacement over 14 h was calculated using Imaris software and reported as the mean ratio of RhoGAP22 mutant-expressing cell displacement to the displacement of cells expressing empty vector. Values are means from three experiments; error bars show standard deviations. Significance was determined by one-way repeated-measures ANOVA. *, P < 0.05.
Ablation of the 14-3-3 binding site affects RhoGAP22 GAP activity.
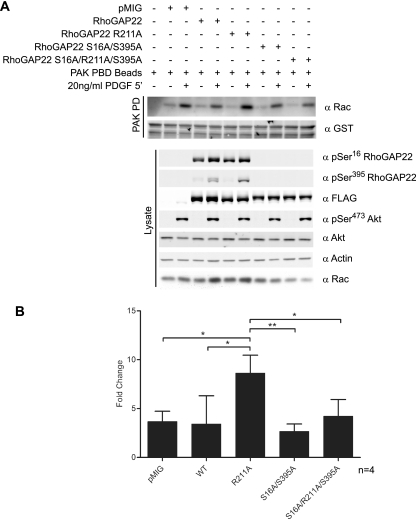
To demonstrate that 14-3-3 binding directly regulated the GAP activity of RhoGAP22, we sought to measure Rac-GTP loading in response to growth factor stimulation using an effector pulldown assay. The p21-activated kinase 1 (PAK1) binds Rac-GTP via its CDC42/Rac interactive binding (CRIB) domain. Producing this fragment of PAK as a GST fusion protein allows Rac-GTP loading to be determined by a GST pulldown assay (5). Using this approach, we assayed Rac-GTP loading in response to PDGF stimulation in NIH 3T3 cells stably expressing RhoGAP22 mutants (Fig. 5A). In response to PDGF, only cells expressing the RhoGAP22 R211A mutant displayed increased Rac-GTP loading compared to control cells (Fig. 5B). Mutation of the 14-3-3 binding site alone (S16A/S395A) had no effect, but in combination with R211A (S16A/R211A/S395A), it reversed the increase in Rac-GTP levels observed with the RhoGAP22 R211A mutant.
Fig. 5.
Mutation of the catalytic arginine of the GAP domain of RhoGAP22 potentiates growth factor-stimulated Rac-GTP loading. (A) NIH 3T3 fibroblasts expressing various RhoGAP22 constructs (described in the text) were serum starved and treated with 20 ng/ml PDGF for 5 min or left untreated. Cells were lysed, subjected to PAK-GST pulldown, and blotted with indicated antibodies. (B) The induction of Rac-GTP loading for each cell line was determined via densitometry. Error bars indicate standard deviations of the means. Significance was determined using Student's unpaired t test. *, P < 0.05; **, P < 0.01.
Knockdown of endogenous RhoGAP22 levels causes defects in insulin-regulated morphological changes.
We next sought to examine the role of endogenous RhoGAP22 in insulin-regulated changes in cell morphology, using an siRNA directed against RhoGAP22. Knockdown of RhoGAP22, but not other Rac-GAPs, had previously been shown to upregulate Rac-GTP loading and alter the mode of cellular motility (27). We obtained siRNA sequences targeting rat RhoGAP22 and were able to achieve a 60% knockdown in RhoGAP22 protein abundance in L6 myoblasts (Fig. 6A). The reduction in RhoGAP22 protein levels caused a defect in insulin-induced cellular morphological change. Changes in cellular morphology were assayed by fixed-cell immunofluorescence microscopy utilizing Alexa 647-conjugated phalloidin to stain the actin cytoskeleton. Measurements of the cell perimeter, cell area, and cellular form factor, a measure of cellular circularity defined as 4πarea/perimeter2, were derived using the Cell Profiler software package (8). Insulin caused a marked increase in circularity in control cells, as defined by an increase in form factor, despite there being no change in cell perimeter or cell area (Fig. 6B and C). In contrast, RhoGAP22 knockdown cells decreased in circularity in response to an insulin stimulus and had a marked increase in cell perimeter compared to control cells (Fig. 6B and C).
DISCUSSION
In this study, we showed that RhoGAP22 is an insulin-responsive and Akt-dependent 14-3-3 binding protein. Further analysis of this interaction yielded three key findings. First, we determined that 14-3-3 binds RhoGAP22 at two phosphoserine residues (pSer16 and pSer395) and that binding of 14-3-3 to the pSer395 residue is dependent on binding to the Ser16 residue. Second, we have demonstrated that Akt is the kinase responsible for phosphorylation of Ser16 in order to mediate 14-3-3 binding to RhoGAP22. Finally, phosphorylation and/or 14-3-3 binding to Rho GAP22 plays an important role in insulin- and growth factor-dependent changes in cell motility by modulating the GAP activity of RhoGAP22. These data lead to a model whereby the interaction of 14-3-3 with RhoGAP22 negatively regulates its GAP activity, modulating the GTP loading of Rac1, likely at a specific location in the cell periphery, leading to localized changes in actin turnover and cell motility.
An intriguing aspect of these data concerns the involvement of Akt as the RhoGAP22 kinase. Phosphorylation of the 14-3-3 binding residues Ser16 and Ser395 is dependent on Akt, and Akt directly phosphorylates one of these sites in vitro. The lack of phosphorylation of the 126-714 S395A construct also indicates that the S476 residue is not an Akt substrate. The fact that Akt phosphorylated only pSer16 yet was required for insulin-stimulated 14-3-3 binding to both sites was interesting. Two models may explain these data. In the first, phosphorylation at Ser395 is dependent on phosphorylation at Ser16. Thus, the absence of Akt activity whether by chemical inhibition or knockout would prevent 14-3-3 binding to Ser16 and thus Ser395. It is possible that the phosphorylation and 14-3-3 binding to the Ser395 site is regulated via a conformational change in RhoGAP22. In this model, RhoGAP22 in the unstimulated state would exist in a closed conformation with the N terminus blocking access to the Ser395 site. As a consequence of insulin or growth factor stimulation, Ser16 would be phosphorylated and bind 14-3-3. Either the phosphorylation event itself or the binding of 14-3-3 would trigger a conformational change, unfolding the protein and allowing access to Ser395 by its kinase, resulting in phosphorylation and 14-3-3 binding (see Fig. S9 in the supplemental material). This model can also explain why inhibition of Akt reduces insulin-stimulated 14-3-3 binding to Ser395 in a RhoGAP22 construct that lacks the Ser16 site (Fig. 3D). In this truncated form of the protein, the entire N terminus, including Ser16, is missing. This would relieve the conformational control on Ser395 phosphorylation and 14-3-3 binding, allowing direct access by its kinase. The one confounding piece of data is the inability of Akt to phosphorylate Ser395 in vitro on a RhoGAP22 fragment that lacks the Ser16 site (Fig. 3C). While Ser395 is predicted in silico to be an Akt site, it is not canonical, as it lacks an arginine residue in the −5 position. It is possible that this is enough to prevent recognition by Akt in vitro, but it may still be recognized in vivo by other protein-protein interactions that recruit Akt to RhoGAP22. The other possible model is that Akt does not directly phosphorylate the Ser395 site but rather this function is regulated by an alternate kinase. If this hypothesis is correct, the alternate kinase is likely downstream of Akt, because inhibition of Akt blocks insulin-stimulated 14-3-3 binding at the Ser395 site (Fig. 3D). We have examined the possible role of a number of putative kinases that might be considered to be downstream of Akt using chemical inhibitors, none of which inhibited insulin-stimulated 14-3-3 binding to the Ser395 site. It was proposed earlier that RhoGAP22 acts to suppress Rac1 activity in response to intracellular signaling through RhoA and its effector ROCK (27), although the authors of that study could find no evidence that ROCK phosphorylates RhoGAP22 directly. Consistent with this, we could find no evidence that 14-3-3 binding to RhoGAP22 is dependent on ROCK activity.
Our GAP activity data strongly imply a role for phosphorylation in the regulation of RhoGAP22 GAP activity. Overexpression of a GAP dead mutant of RhoGAP22 (R211A) markedly potentiated GTP loading of Rac but only in the presence of growth factor. The most likely interpretation of these data is that growth factor stimulation inactivates the GAP activity of RhoGAP22 and that the R211A mutant sequesters GTP-loaded Rac, protecting it from other GAPs/effectors. The potentiation observed in the presence of growth factor is likely due to an effect of the growth factor on Rac GEF activity (14), which in itself increases the level of Rac-GTP. Intriguingly, despite this increase in Rac-GTP loading in cells overexpressing the R211A mutant, we did not observe any effect of this mutant on cell motility, presumably because other factors further downstream likely become rate limiting in this situation. The fact that this phenomenon was not observed when the phosphorylation sites in the R211A mutant were disarmed (S16A/R211A/S395A) likely invokes an alternate function, possibly involving the localization of the GAP to a particular site in the cell, where it can associate with growth factor-activated Rac. It is interesting that different pools of Rac have been identified at the leading and trailing edges of the cell, with the majority of Rac-GTP being localized to the leading edge. Interestingly, the S16A/S395A mutant significantly impaired cell motility, possibly consistent with the fact that this protein is a mislocalized constitutively active Rac GAP that disrupts the organization of Rac-GTP that is required for directional movement. Due to the precise spatiotemporal regulation of this process, an effect on Rac-GTP loading was unlikely to be seen in vivo (Fig. 5). Consistent with this model, when the GAP activity of this mutant was disarmed (S16A/R211A/S395A), no significant effect on cell motility was observed (Fig. 4).
Rho GAP22 is widely expressed in a variety of mammalian cell types (Fig. 1E) and plays an important role in cell migration (27). Indeed, the transition from mesenchymal and amoeboid modes of cell migration, which RhoGAP22 regulates, plays a critical role in cancer cell metastasis (29). Insulin and growth factors such as PDGF have previously been shown to promote cell migration and membrane ruffling, with both of these processes being dependent on Rac activity (9). Consistent with a role in the regulation of Rac-GTP loading, siRNA-mediated knockdown of endogenous RhoGAP22 caused aberrant cell morphology in insulin-stimulated cells (Fig. 6). Knockdown of RhoGAP22 has been shown to increase total cellular Rac-GTP levels (27). An increase in Rac-GTP levels is known to cause cell spreading (26), and so the aberrant morphological changes we observed were therefore probably due to the formation of multiple lamellipodia, resulting in a loss in cell roundness. Akt is a well-known oncogene and has previously been implicated in the regulation of cell motility (10, 11, 23, 30), upstream of Rac1 activation (13, 15). However, it has not previously been demonstrated that Akt can directly control the activity of Rac by signaling through a GAP. Thus, RhoGAP22 provides a strong link between insulin- and growth factor-mediated Akt signaling and the control of Rac. In order for directed migration to occur, Rac activity must be subjected to precise regulation. Even a moderate increase in Rac1-GTP levels promotes more random migration, via the generation of multiple competing lamellipodia (12). RhoGAP22 could therefore restrict Rac activity to allow directed cell migration in response to Akt signaling, and perturbations in this pathway may play important roles in cancer metastasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Burchfield and Katarina Mele for assistance with microscopy and image analysis, respectively. Mouse p68RacGAP cDNA was a gift from Cam Patterson from the University of North Carolina. Double Akt knockout MEFs were a gift from Morris Birnbaum at University of Pennsylvania. CHO IR/IRS-1 cells were a gift from Morris White, Harvard Medical School. Plat-E cells were a gift from T. Kitamura from University of Tokyo. Mass spectrometric analysis for this work was carried out at the Bioanalytical Mass Spectrometry Facility, University of New South Wales (UNSW).
Mass spectrometric analysis for this work was supported in part by grants from the Australian Government Systemic Infrastructure Initiative and Major National Research Facilities Program (UNSW node of the Australian Proteome Analysis Facility) and by the UNSW Capital Grants Scheme. This work was supported by grants from the National Health and Medical Research Council of Australia and Diabetes Australia Research Trust (to D.E.J.). D.E.J. is an NHMRC senior principle research fellow.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Abe K., et al. 2000. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 275:10141–10149 [DOI] [PubMed] [Google Scholar]
- 2. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with Image J. Biophotonics Int. 11:36–42 [Google Scholar]
- 3. Aitsebaomo J., et al. 2004. p68RacGAP is a novel GTPase-activating protein that interacts with vascular endothelial zinc finger-1 and modulates endothelial cell capillary formation. J. Biol. Chem. 279:17963–17972 [DOI] [PubMed] [Google Scholar]
- 4. Arrieumerlou C., Meyer T. 2005. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev. Cell 8:215–227 [DOI] [PubMed] [Google Scholar]
- 5. Azim A. C., Barkalow K., Chou J., Hartwig J. H. 2000. Activation of the small GTPases, rac and cdc42, after ligation of the platelet PAR-1 receptor. Blood 95:959–964 [PubMed] [Google Scholar]
- 6. Bae S. S., Cho H., Mu J., Birnbaum M. J. 2003. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J. Biol. Chem. 278:49530–49536 [DOI] [PubMed] [Google Scholar]
- 7. Brummer T., et al. 2008. Phosphorylation-dependent binding of 14-3-3 terminates signalling by the Gab2 docking protein. EMBO J. 27:2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpenter A. E., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheresh D. A., Leng J., Klemke R. L. 1999. Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J. Cell Biol. 146:1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung C. Y., Potikyan G., Firtel R. A. 2001. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7:937–947 [DOI] [PubMed] [Google Scholar]
- 11. Enomoto A., et al. 2005. Akt/PKB regulates actin organization and cell motility via girdin/APE. Dev. Cell 9:389–402 [DOI] [PubMed] [Google Scholar]
- 12. Gail M. 1973. Locomotion of tissue cells. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 13. Genot E. M., et al. 2000. The T-cell receptor regulates Akt (protein kinase B) via a pathway involving Rac1 and phosphatidylinositide 3-kinase. Mol. Cell Biol. 20:5469–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han J., et al. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279:558–560 [DOI] [PubMed] [Google Scholar]
- 15. Higuchi M., Masuyama N., Fukui Y., Suzuki A., Gotoh Y. 2001. Akt mediates Rac/Cdc42-regulated cell motility in growth factor-stimulated cells and in invasive PTEN knockout cells. Curr. Biol. 11:1958–1962 [DOI] [PubMed] [Google Scholar]
- 16. Kolsch V., Charest P. G., Firtel R. A. 2008. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 121:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larance M., et al. 2010. Global phosphoproteomics identifies a major role for Akt and 14-3-3 in regulating EDC3. Mol. Cell. Proteomics 9:682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Letunic I., Doerks T., Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linding R., Russell R. B., Neduva V., Gibson T. J. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morita S., Kojima T., Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1069 [DOI] [PubMed] [Google Scholar]
- 21. Obata T., et al. 2000. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275:36108–36115 [DOI] [PubMed] [Google Scholar]
- 22. Obenauer J. C., Cantley L. C., Yaffe M. B. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Primo L., et al. 2007. Essential role of PDK1 in regulating endothelial cell migration. J. Cell Biol. 176:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puntervoll P., et al. 2003. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31:3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramm G., Larance M., Guilhaus M., James D. E. 2006. A role for 14-3-3 in insulin-stimulated GLUT4 translocation through its Interaction with the RabGAP AS160. J. Biol. Chem. 281:29174–29180 [DOI] [PubMed] [Google Scholar]
- 26. Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanz-Moreno V., et al. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell 135:510–523 [DOI] [PubMed] [Google Scholar]
- 28. Scholz R.-P., et al. 2009. DLC1 interacts with 14-3-3 proteins to inhibit RhoGAP activity and block nucleocytoplasmic shuttling. J. Cell Sci. 122:92–102 [DOI] [PubMed] [Google Scholar]
- 29. Wolf K., et al. 2003. Compensation mechanism in tumor cell migration. J. Cell Biol. 160:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoeli-Lerner M., et al. 2005. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol. Cell 20:539–550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.