Table 1.
Properties of 3T3sw mitochondrial nucleoids, mitochondria, and relative DNA packing density
| Property | Value | Basis or reference |
|---|---|---|
| Mean no. of nucleoids/3T3sw cell (±SD)d | ||
| PicoGreen DNA staining | 275 (±77) | |
| TFAM-mEos2 (green state) labeling | 291 (±92) | |
| Mean no. of mtDNA copies/3T3sw cellb (±SD) | 821 (±163) | |
| mtDNA/nucleoidsc | 2.98 | 821/275 |
| Mean nucleoid vol (nm3) | 830,000 | Fig. 4 |
| Mean matrix space between cristae (nm)d (±SD) | 68 (±24) | |
| Mean matrix diam (nm) (±SD)e | 238 (±75) | |
| ρpackf | ||
| Bacteriophage T7 | 0.49 | 48 |
| ECV304 (human) mt nucleoid | 0.110 | 24 |
| P. polycephalum mt nucleoid | 0.069 | 5 |
| 3T3 (mouse) mt nucleoid | 0.063g | |
| Nuclear DNA | 0.031 | 3.3 × 109 bp/113 μm3 |
| E. coli | 0.005 | 4.64 × 106 bp/0.9 μm3 |
| Nucleoid diam (nm) | ||
| 2D PALM (TFAM-mEos2) | 110 (±59) | |
| dSTORM (DNA antibody) | 110 (±46) |
Data were determined by whole-cell confocal microscopy.
Data were determined by qPCR.
Average mtDNA genomes/cell divided by average nucleoids/cell.
Data were determined by measurements from EM images using the formula (mitochondrial length/number of cristae) − average crista width (21.5 nm); n = 80 mitochondria.
Data were determined from 60 width measurements taken between inner boundary membranes; sample range, 115 to 430 nm.
Data were determined using the equation
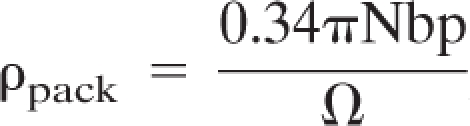 , where Nbp represents the number of base pairs and Ω represents the package volume in cubic nanometers. mt, mitochondrial.
, where Nbp represents the number of base pairs and Ω represents the package volume in cubic nanometers. mt, mitochondrial.
Data were determined using averages of 3 genomes/nucleoid and the equation shown in footnote f.
