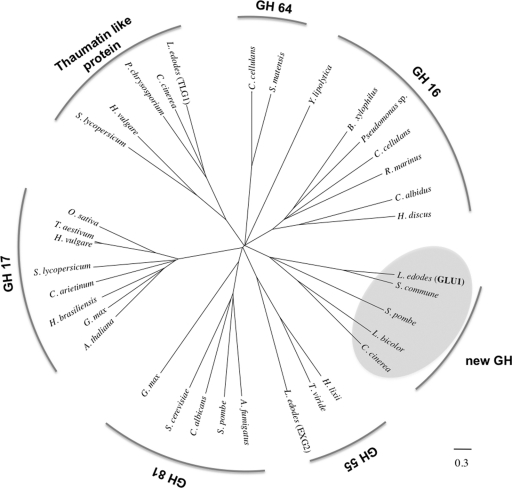
Fig. 3.
Phylogenetic relationship of L. edodes glucanases (GLU1, TLG1, and EXG2), thaumatin-like proteins, and EC 3.2.1.39 β-1,3-glucanases on the CAZy server. Sequences of GLU1, TLG1, EXG2, fungal proteins having homology with GLU1 (XP_003029493 from Schizophyllum commune, NP_592836 from Schizosaccharomyces pombe, XP_001873506 from Laccaria bicolor, and XP_001829384 from Coprinopsis cinerea, highlighted by gray circles), thaumatin-like proteins (CAA50059 from Solanum lycopersicum, AAK55325 from Hordeum vulgare, genome database of Phanerochaete chrysosporium, and XP_001837765 from Coprinopsis cinerea), GH family 16 members (BAE02683 from Bursaphelenchus xylophilus, AAZ04385 from Chlamys albidus, BAH84971 from Haliotis discus, XP_500934 from Yarrowia lipolytica, AAC44371 from Cellulosimicrobium cellulans, BAC16331 from Pseudomonas sp., and AAC69707 from Rhodothermus marinus), GH family 17 members (AAA32756 from Arabidopsis thaliana, CAA10167 from Cicer arietinum, AAA33946 from Glycine max, CAB38443 from Hevea brasiliensis, AAA32958 from Hordeum vulgare, AAD10382 from Oryza sativa, AAA03617 from Solanum lycopersicum, and CAA77085 from Triticum aestivum), GH family 55 members (CAA58889 from Hypocrea lixii and BAD67019 from Trichoderma viride), GH family 64 members (AAA25520 from Cellulosimicrobium cellulans and BAA34349 from Streptomyces matensis), and GH family 81 members (AF121133 from Aspergillus fumigatus, CAB62580 from Candida albicans, BAA11407 from Glycine max, AAB82378 from Saccharomyces cerevisiae, and CAA91245 from Schizosaccharomyces pombe) were aligned using MAFFT (version 6; http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) with the E-INS-I algorithm. A phylogenetic tree was inferred from the alignments with the minimum linkage method and drawn by using FigTree software (version 1.1.2; http://tree.bio.ed.ac.uk/software/figtree/).