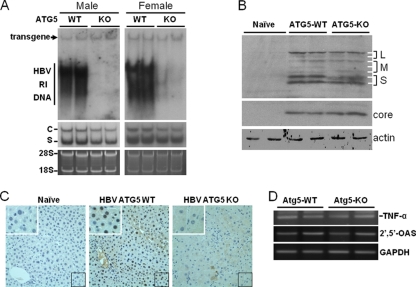
Fig. 4.
Analysis of HBV replication in mouse livers. (A) Southern (top panel) and Northern (middle panel) blot analyses. For the isolation of HBV RI DNA, mouse livers were homogenized in the radioimmunoprecipitation assay (RIPA) solution (10 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate), treated with proteinase K, and phenol extracted using our previous procedures (21). The DNA was then subjected to Southern blot analysis using 32P-labeled HBV DNA as the probe. The HBV transgene served as the loading control. For the isolation of total RNA, mouse livers were homogenized in TRIzol (Invitrogen), and RNA was isolated as previously described (21). C and S denote the HBV C gene and S gene transcripts, respectively. The 28S and 18S ribosomal RNAs were used as the loading controls for Northern blot analysis (bottom panel). Left panels, male mice; right panels, female mice. (B) Western blot analysis of HBsAg (upper panel) and HBV core protein (lower panel). Liver homogenates as mentioned above were subjected to Western blot analysis. The α-actin protein was also analyzed to serve as the loading control. L, M, and S denote the locations of large, middle, and small HBsAg proteins. The two L protein bands and the two S protein bands represent the glycosylated and the nonglycosylated forms of their respective protein species. The three M protein bands represent doubly glycosylated, monoglycosylated, and nonglycosylated M protein forms, as previously reported (15, 19). (C) Immunohistochemistry analysis of the HBV core protein. Liver tissue sections were stained with the mouse anti-HBcAg antibody (Abcam) and the alkaline phosphatase-conjugated rabbit anti-mouse secondary antibody. A naïve mouse without HBV was used as the control in the staining experiment. The area boxed in the lower right corner of individual panels was enlarged and is shown in the upper left corner of that particular panel. (D) Semiquantitative reverse transcription (RT)-PCR analysis of TNF-α and 2′,5′-OAS RNAs in the mouse liver. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA was also analyzed to serve as a control. Details of the semiquantitative RT-PCR analysis for the RNAs of 2′,5′-OAS and GAPDH have been described previously (21). The forward and reverse primers used for the TNF-α RT-PCR analysis were 434-CCCACGTCGTAGCAAACCAC-453 and 610-CGTAGTCGGGGCAGCCTTGTC-590, respectively.