Abstract
The influenza virus transcribes and replicates its genome inside the nucleus of infected cells. Both activities are performed by the viral RNA-dependent RNA polymerase that is composed of the three subunits PA, PB1, and PB2, and recent studies have shown that it requires host cell factors to transcribe and replicate the viral genome. To identify these cellular partners, we generated a comprehensive physical interaction map between each polymerase subunit and the host cellular proteome. A total of 109 human interactors were identified by yeast two-hybrid screens, whereas 90 were retrieved by literature mining. We built the FluPol interactome network composed of the influenza virus polymerase (PA, PB1, and PB2) and the nucleoprotein NP and 234 human proteins that are connected through 279 viral-cellular protein interactions. Analysis of this interactome map revealed enriched cellular functions associated with the influenza virus polymerase, including host factors involved in RNA polymerase II-dependent transcription and mRNA processing. We confirmed that eight influenza virus polymerase-interacting proteins are required for virus replication and transcriptional activity of the viral polymerase. These are involved in cellular transcription (C14orf166, COPS5, MNAT1, NMI, and POLR2A), translation (EIF3S6IP), nuclear transport (NUP54), and DNA repair (FANCG). Conversely, we identified PRKRA, which acts as an inhibitor of the viral polymerase transcriptional activity and thus is required for the cellular antiviral response.
INTRODUCTION
Influenza A virus is responsible for annual epidemics of respiratory disease and reoccurring pandemics and represents an important public health problem worldwide. Its genome is composed of eight single-stranded negative-polarity viral RNA (vRNA) segments. They are individually encapsidated by nucleoprotein (NP) and the RNA-dependent RNA polymerase (RdRP), forming a viral ribonucleoprotein (vRNP) complex (reviewed in reference 38). Each vRNA behaves as an independent template for transcription and replication that are both taking place in the nucleus of infected cells. The virus RNA polymerase is a heterotrimer composed of subunits PA, PB1, and PB2. PB1 is the core subunit of the complex and contains the polymerase activity, while PB2 recognizes capped cellular mRNA (38) and PA possesses an endonuclease activity (12). After viral decapsidation, vRNPs are transported into the nucleus, where they are engaged in primary transcription. This is initiated by cap snatching of cellular pre-mRNA: the PB2 subunit recognizes capped mRNAs, while the PA subunit cleaves them 10 to 15 nucleotides (nt) downstream of the cap, generating cap-containing primers for virus mRNA synthesis (28). Viral mRNAs are then translated by cytoplasmic ribosomes, allowing newly synthesized components of the viral polymerase and NP to accumulate in the nucleus. It has recently been proposed that the nuclear accumulation of newly synthesized viral proteins could be responsible for the switch from viral transcription to replication (56). For the genome replication, untranslated sequences at the 5′ and the 3′ ends of each genomic vRNA segment act as promoter elements that are recognized by the viral polymerase. vRNA segments are copied into positive-strand complementary RNAs (cRNAs), which are encapsidated by NP and serve as templates for de novo vRNA synthesis (22, 38).
The influenza virus polymerase performs numerous functions during the virus life cycle, suggesting that many cellular factors interact with this complex and are required for the viral genome's transcription and replication.
Only a few cellular partners have been previously described in the literature, but recent studies identified several influenza virus polymerase interactors using proteomic approaches (23, 33). Moreover, a global survey of influenza virus host cell partners was established using the yeast two-hybrid (Y2H) technology (51). A series of functional genome-wide small interfering RNA (siRNA) screenings were also conducted for the identification of host factors involved in influenza virus replication (5, 18, 24, 27, 51; reviewed in reference 58). We present here a more specific analysis that is focused on the RdRP cellular interactors. We reconstructed a global influenza virus polymerase physical interaction map by performing yeast two-hybrid screens with each subunit as bait and by retrieving protein-protein interactions from the literature. Through this interaction network, we identified cellular functions specifically targeted by the viral polymerase. We selected nine functionally relevant host cell partners, whose interactions with the viral proteins were validated in human cells. We show that these cellular factors affect both the viral polymerase activity and the virus replication using functional assays: eight of them are required for virus replication and polymerase activity. Conversely, PRKRA acts as an antiviral factor since virus replication and polymerase activity are enhanced when its expression is depleted.
MATERIALS AND METHODS
Cloning of influenza virus ORFs.
NP, PA, PB1, and PB2 open reading frames (ORFs) from A/PR/8/34 (H1N1) and A/WSN/33 (H1N1) (kindly provided by G. Brownlee and V. Moulès, respectively), PA and PB1 from A/VietNam/1194/2004 (H5N1) (kindly provided by V. Moulès), PA from A/Turkey/651242/2006 (H5N1) (the H5N1 genomic RNAs were kindly provided by V. Moulès), and PB2 from A/Victoria/3/75 (H3N2) (kindly provided by D. Hart) were amplified from plasmids encoding corresponding cDNA genomic segments or from genomic viral RNA by using ORF-specific Gateway primers (containing attB1.1 at the 5′ end and attB2.1 at the 3′ end and without ATG and stop codons [48]). Four PB2 fragments isolated from the ESPRIT technology were screened: the long (amino acids [aa] 234 to 496) and the short (aa 318 to 483) (17) cap-binding domains, the “627” domain (aa 538 to 693), encompassing the K627 residue (55), and the C-terminus domain (CTD) (DPDE; aa 678 to 759) (54). We constructed the following compensatory point mutation mutants: K627E in the 627 domain (52) and D701N (49), R702K (15), and the D701N R702K double-point mutant in the DPDE domain. After PCR, ORFs were cloned by in vitro recombination into donor vectors (pDONR207/223). All clones were sequence verified and stored in the viralORFeotheque repository. We developed a database that provides an integrated set of bioinformatic tools to clone viral ORFs in the Gateway system, viralORFeome (http://www.viralorfeome.com [43]).
Y2H assay.
Viral ORFs were cloned by recombination into pGBKT7-gw and transformed in AH109 (bait strain; Clontech). Y2H screens were performed by yeast mating (53). Briefly, the human cDNA libraries (comprising spleen, fetal brain, and respiratory epithelium libraries from Invitrogen and customized with CloneMiner for the latter) were transformed in Y187 (prey strain; Clontech), and each bait strain was mated with the library prey strain. Diploids were plated on synthetic defined medium lacking tryptophan, leucine, and histidine and supplemented with 3-aminotriazole (SD−W−L−H +3-AT), and positive clones were streaked twice on this selection medium. AD-cDNAs (where AD refers to the Gal4 activation domain fused to cDNA) were PCR amplified and sequenced. Interaction sequence tags were analyzed through pISTil and deposited in the viralORFeome database (42). All partners of PB2 domains identified in Y2H screens were retested against each PB2 fragment in a Y2H array system (in an all-against-all matrix). First, AD-cDNAs encoding cellular interactors identified with PB2 domains were transformed in the yeast prey strain together with linearized pACT2-gw. Bait and prey strains were arrayed in a 96-well format using a robotic workstation (Tecan Freedom Evo) and mated in an all-against-all array on YPD plates. Diploids were selected on SD −W −L for 2 days, and then transferred on selection medium (SD −W −L −H plus increasing concentrations of 3-AT). Interactions were scored as positive if observed at least twice in 3 independent arrays (53). Among the 79 partners identified with PB2 domains, 59 were positively scored, confirming the results obtained during the screen. The 20 remaining partners were discarded.
GO category enrichment using BiNGO and Golorize on Cytoscape.
BiNGO (Biological Network Gene Ontology) is a Cytoscape plugin that assesses which Gene Ontology (GO) categories are overrepresented in a network (32). It provides single P values, calculated with the hypergeometric test, and takes into consideration both the total number of genes from the analyzed data set and the total number of genes that are linked to the same ontology term, as well as multiple testing of corrected P values, calculated using the Benjamini and Hochberg false discovery rate (FDR) correction, for the enrichment of each GO term (in our case, “biological process”). GOlorize is a Cytoscape plugin that highlights the class members of the enriched categories identified by BiNGO using a color code within the Cytoscape-built network (16, 50). The cross-validation of our Y2H interaction data set revealed a false-positive rate of 15%. To take into account of this rate in the functional enrichment analysis, we generated 50 different clusters of 93 randomly chosen Y2H interactors (representing 85% of the 109 Y2H interactors) plus all of the cellular partners in the literature. For each cluster, we performed a GO analysis using BiNGO. We considered positive the overrepresented classes that were scored positive for all the 50 clusters. Nine functional categories were selected, and the class members were colorized within the FluPol network using GOlorize. Note that we did not change the network layout after the GOlorize analysis.
FluPol network visualization.
We used Cytoscape Web API for visualizing and manipulating the FluPol network graph, allowing a dynamic network display (31; http://flupol.lyon.inserm.fr). All of the interaction data can be exported through this web page.
BiFC.
For the bifunctional fluorescence complementation (BiFC) assay, A/WSN/33 PA, PB1, and PB2 were cloned into Polyc vector, where they were fused upstream of the C-terminal moiety of yellow fluorescent protein (YFP). The cellular partners' genes were amplified from the human cDNA Y2H libraries and then in vitro cloned into pDONR207 as viral ORFs (see above), or were retrieved from the hORFeome collection (48). They were then cloned by recombination in pGWEN, in fusion downstream of the N-terminal moiety of Venus (modified YFP) (26). HEK-293T cells were cotransfected using JetPei (Polyplus) with each combination of plasmid pair in a 96-well plate. Forty-eight hours after transfection, cells were resuspended in 100 μl phosphate-buffered saline (PBS), and YFP signal was measured using a FACSArray (BD). The percentage of fluorescent cells was measured (mean of three independent experiments), and an interaction was considered positive when more than 5% of cells were fluorescent, according to results obtained with negative controls.
FCPI assay.
The flow cytometry protein interaction (FCPI) assay was set up using two couples of interacting proteins (see Fig. S1 in the supplemental material). Viral ORFs (A/WSN/33 strain) were cloned by in vitro recombination into pCMV-BioEase-Cherry-gw, where they are fused downstream of the BioEase tag (Invitrogen) and the fluorescent protein mCherry. Cellular genes were transferred to pCMV-eGFP-gw (kindly provided by Y. Jacob), to be fused downstream to enhanced green fluorescent protein (EGFP). Human HEK-293T cells were cotransfected using JetPei (Polyplus) with each combination of plasmid pair (1 μg of each vector) in a 12-well plate and lysed 48 h after transfection. Sixty micrograms of protein extracts was suspended in 200 μl lysis buffer (150 mM NaCl, 20 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% NP-40 plus Complete miniprotease inhibitor cocktail [Roche]) containing 1 μl of streptavidin-conjugated microspheres (Polysciences) on a multiwell filter plate (Pall) and incubated for 2 h at 4°C under gentle agitation. Each affinity purification was performed in triplicate. The beads were washed twice with 200 μl of wash buffer (same as lysis buffer, without the protease inhibitor cocktail) using a vacuum manifold and then were resuspended in 100 μl of wash buffer, and bead-associated GFP signal was analyzed using a FACSArray (BD). The analysis of results is based on the relative mean fluorescence intensity (MFI) associated with the beads compared to the MFI of an empty bait vector. An interaction is considered positive when the difference is higher than 5%.
Cells and viruses.
A549 and HEK-293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 1% penicillin/streptomycin and 10% fetal bovine serum at 37°C and 5% CO2. Influenza A virus A/New Caledonia/2006 (H1N1) (clinical isolate) was propagated in MDCK cells. Titers of viral stocks were determined by PFU assay on MDCK cells.
Coimmunoprecipitation and protein detection.
Cellular genes were transferred to pCIneo-3Flag-gw (kindly provided by Y. Jacob), to be fused downstream to 3× Flag epitope (Sigma). A549 cells were transfected in 6-well plates using Turbofect (Fermentas). Twenty-four hours after transfection, cells were infected by H1N1 A/New Caledonia/2006 virus at a multiplicity of infection (MOI) of 1 and lysed 24 h after infection. Three hundred micrograms of protein extracts was suspended in 1 ml lysis buffer containing 20 μl of anti-Flag M2 magnetic beads (Sigma) and incubated for 2 h at 4°C under gentle agitation. The beads were washed three times with 1 ml of wash buffer on a magnetic rack, and then precipitates were eluted in 20 μl SDS loading buffer without reducing agents and boiled for 5 min at 95°C. Total cell extracts (20 μg) and immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting using antibodies against PB1 (rabbit polyclonal antibody kindly provided by J. Ortin) and Flag (horseradish peroxidase [HRP]-conjugated mouse monoclonal antibody; Sigma). The secondary anti-rabbit antibody was also HRP conjugated (Santa Cruz). EGFP-PRKRA was detected with mouse monoclonal anti-GFP antibody (Sigma).
Influenza virus replication assay and virus titer determination using MUNANA.
Human A549 cells were transfected with siRNA targeting each gene (using a pool of 2 siRNAs at a concentration of 40 nM; Invitrogen) in a reverse transfection procedure with Lipofectamine RNAiMAX (Invitrogen) in a 96-well plate. Forty-eight hours after siRNA transfection, cells were infected by H1N1 A/New Caledonia/2006 virus at an MOI of 0.5 in DMEM containing 1% penicillin/streptomycin and 0.25 μg/ml TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (Sigma). Cell culture supernatants (25 μl) were collected at 24 h and 48 h postinfection, and virus titers were determined by quantifying the neuraminidase activity using a MUNANA [2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid] assay, as described in reference 59, with minor modifications. Briefly, cell supernatants were transferred into a 96-well black flat-bottom plate and mixed with 25 μl of PBS with Ca2+/Mg2+ (Invitrogen) and 50 μl of MUNANA stock solution (20 μM; Sigma). Plates were incubated 1 h at 37°C, and the reaction was stopped by adding 100 μl of Stop solution (glycine 0.1 M [pH 10.7], 25% ethanol). The amount of fluorescent product released by MUNANA hydrolysis (4-methylumbelliferyl [4-MU]) was measured in a Tecan spectrophotometer with excitation and emission wavelengths of 365 and 450 nm, respectively. A reference curve was established in parallel with serial dilutions of a titrated stock of influenza virus.
To assess potential cellular toxicity induced by siRNAs, a cell viability assay was performed using the resazurin-based fluorometric assay at 48 h posttransfection (see Fig. 2C). Resazurin (1 mg/ml of medium; Sigma) detects cell viability by converting to a red fluorescent dye in response to reduction of growth medium resulting from cell growth. The fluorescent signal generated is proportional to the number of living cells. After an incubation for 2 h at 37°C, the fluorescence was measured with a Tecan spectrophotometer with excitation and emission wavelengths of 550 and 590 nm, respectively. A reference curve was established with known serial dilutions of growing cells.
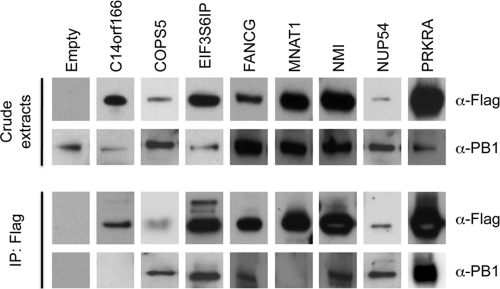
Fig. 2.
Interaction of the influenza virus polymerase and host cell partners in infected cells. A549 cells were transfected with the indicated proteins fused to 3× Flag. Twenty-four hours after transfection, cells were infected with H1N1 A/New Caledonia/2006 at an MOI of 1. Cells were lysed 24 h after infection, and protein lysates were immunoprecipitated (IP) with anti-Flag (α-Flag) magnetic beads. Total cell extracts and immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting using antibodies against PB1 (α-PB1) and Flag.
Influenza virus minigenome replicon assay. (i) Overexpression of human partner proteins.
Human HEK-293T cells (plated in a 96-well plate) were cotransfected using JetPei (Polyplus) with a combination of plasmids expressing a cellular partner (100 ng), vectors encoding the A/WSN/33 polymerase subunits (PA, PB1, and PB2) and NP (50 ng each plasmid), a reporter plasmid (pPOLI-Luc-RT, encoding the firefly luciferase in the negative-sense orientation flanked by the noncoding regions of the segment 8 of A/WSN/33, driven by a polymerase I [Pol I] promoter, kindly provided by M. Shaw [19]; 50 ng), and a vector constitutively expressing Renilla luciferase (5 ng pRL-SV40-Rluc; Promega). Twenty-four hours and 48 h after transfection, luciferase activities were determined using the Dual-Glo luciferase assay system (Promega). As a positive control, cells were transfected by the complete set of vRNP and an empty cellular protein expression vector. As a negative control, cells were transfected with the same reaction mixture but without the plasmid expressing PB1.
(ii) Depletion of human partner genes.
Human HEK-293T cells were transfected with siRNA targeting each gene as described above. Knockdown was allowed to proceed for 48 h, and then cells were transfected with the complete set of vRNP components (plasmids expressing NP, PA, PB1, and PB2 and the reporter vector encoding firefly luciferase) as described above. As positive control, cells were transfected by a scrambled control siRNA, and then with the complete set of the vRNP components. As a negative control, cells were treated with the same mixture but without PB1-expressing plasmid.
RESULTS AND DISCUSSION
Building an influenza virus polymerase cellular interactome network.
We generated a comprehensive protein-protein interaction network between the viral influenza A virus polymerase (FluPol) and its host cell partners using the yeast two-hybrid (Y2H) system. For this purpose, human cDNA libraries were screened with the RdRP subunits and NP from different virus strains (H1N1, H3N2, and H5N1). PB2 fragments that were isolated by the ESPRIT technology and of which structures were defined were also screened individually (17, 54, 55, 60). Human cDNA libraries from three different tissues (spleen, fetal brain, and respiratory epithelium) were used, covering a wide range of the human proteome as well as a tissue that is specifically targeted by the influenza A virus. Altogether, we discovered 112 viral-human protein interactions involving 109 human proteins, including 5 interactions with NP, 30 with PA, 18 with PB1, and 59 with PB2. Among them, 3 human proteins interact both with PA and PB2 (see Table S1 in the supplemental material and Fig. 1A and 1B).
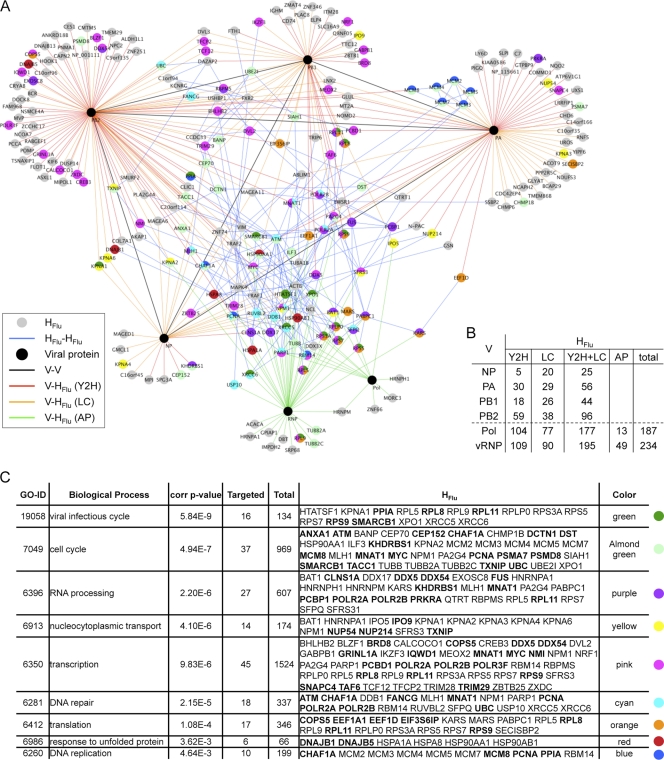
Fig. 1.
The influenza virus polymerase (FluPol) interaction network. (A) Graphical representation of the FluPol interactome map. Black nodes represent viral proteins. V represents NP, PA, PB1, and PB2. Pol corresponds to the polymerase complex, and RNP to the ribonucleoprotein particle, composed of all 4 proteins and vRNA. Gray nodes represent human proteins (HFlu). Red, orange, and green edges represent interactions between viral and human proteins: V-HFlu, identified by the Y2H assay (I-MAP data set) is in red, literature mining is in orange, and affinity purification is in green. Black edges represent interactions between viral proteins (V-V), and blue edges represent interactions between human proteins (HFlu-HFlu). The FluPol interaction network is available online; for a better visualization and dynamic display, see http://flupol.lyon.inserm.fr. Human proteins of enriched classes are colored according to the table in panel C. (B) Number of FluPol interactors. Data are given for the Y2H data set, literature curated interactions (LC), and the affinity purification (AP) approach by viral protein, the polymerase, and the vRNP (for the AP approaches). (C) Table of the overrepresented classes (GO biological process) in the FluPol interactome. The corrected P value column corresponds to a Benjamini and Hochberg FDR correction. Also indicated are the number of targeted proteins (which are detailed in the HFlu column) and the total proteins within each class. A color was assigned to each overrepresented class, allowing their positioning on the interactome map.
To assess the confidence of the yeast two-hybrid results, two validation methods in human cells were used. Thirty-four cellular partners (31% of the Y2H interactors) were randomly retested for their interaction with the viral protein bait that trapped them in the Y2H assay by either bifunctional fluorescence complementation (BiFC) (26) or a flow cytometric method developed in our lab to measure protein-protein interactions (FCPIA, similar to the assay developed by the Neubig lab [4]). While the bait (in our case, the viral protein) is fused to both the BioEase tag, an in vivo biotinylated sequence that can be trapped using streptavidin beads, and to the fluorescent protein mCherry (allowing its detection on beads), the prey is fused to eGFP. This assay allows detection of copurified preys on streptavidin beads using a flow cytometer (Materials and Methods; see Fig. S1 in the supplemental material). KPNA1 was used as positive control as it interacts with PB2 and NP (39, 54). Among the 34 host cell partners tested, 29 were validated using either BiFC, FCPIA, or both assays (in green in Table 1; see Table S5 in the supplemental material). We identified 18 new connections through these validation assays, by performing an array in which the cellular proteins were a priori tested against different viral baits (in blue in Table 1). It is of note that we did not validated the interactions identified in the Y2H assay for three cell partners but discovered interactions with different viral proteins (EIF3S6IP, FANCG, and ZCCHC17). Twenty-nine of the 34 Y2H interactors in the subset tested were validated, giving a false-positive rate of 15%, which is in accordance with previous studies (9). Most of the cellular proteins interact with one FluPol subunit (17 of the 29 validated), like C14orf166, which only interacts with PA, or EEF1A1, which only interacts with PB1. We also observed that a few host cell partners interact with both PB1 and PB2 (the 5 cellular proteins CES1, CHAF1A, DDX54, MNAT1, and NMI). PCNA and PSMA7 interact with the FluPol subunits (PA, PB1, and PB2), whereas EEF1D interacts with all of the vRNP protein constituents.
Table 1.
List of consolidated FluPol interactorsa
Green, interaction found in Y2H and validated in either BiFC or FCPIA; yellow, interaction that was tested either with BiFC or FCPIA but was negative; blue, novel interaction detected in either BiFC or FCPIA. Raw values are given in Table S5 in the supplemental material.
The peGFP-empty vector (eGFP/) was used as a negative control in FCPIA.
The pGwen-empty vector (Gwen/) was used as a negative control in BiFC.
We also retrieved virus-host protein interactions from the literature and added them to the VirHostNet knowledge base (37). In total, 26 interactions were identified involving 23 human proteins. (All of the papers describing individual interactions can be retrieved using Table S1 in the supplemental material, where we have indicated the PubMed ID number.) In addition, Shapira et al. identified recently 90 interactions involving 69 cellular proteins in their Y2H screens with the viral polymerase subunits (51). Moreover, we added 49 cellular proteins that were identified as vRNP or polymerase complex host cell partners by proteomic approaches, even though we don't know precisely with which viral protein they actually interact (23, 33). The overlap between these four data sets is low, but not surprising (Table 2). (Note that these data sets include the Y2H set from this report, which is further referred to as the “I-MAP” [for “Infection Mapping Project”] data set, the Y2H set from reference 51, and the literature curated [LC] and proteomics [affinity purification; AP] sets.) In a previous work, where we performed a comprehensive analysis of the hepatitis C virus (HCV) interaction network, we obtained such results: among 278 human proteins interacting with HCV proteins identified in the Y2H set, only 10 were already described in the literature, representing 3.6% overlap (9). This is due to the different methods that were used (e.g., the Y2H assay for I-MAP versus affinity purification-mass spectometry [MS] for the AP set, representing 1.8% overlap). The low overlap (0.9%) between our results and those from Shapira et al. (51), although both studies were conducted using the Y2H assay, could be explained by different procedures that were used, including the following: different libraries (a normalized library of 12,000 full-length ORFs for the study by Shapira et al. versus 3 human cDNA libraries encompassing a large fraction of the human proteome, but including many truncated coding sequences for I-MAP), different screening techniques (mating of a minipool of prey strains for the study by Shapira et al. versus mating of one bait strain against a whole library of prey strains for I-MAP), different Y2H strains (Y8800 and Y8930 for the study by Shapira et al. versus AH109 and Y187 for I-MAP), and different reporter genes (ADE2 and HIS3 for the study by Shapira et al. versus HIS3 for I-MAP) (51). This is supported by a comprehensive interactome analysis conducted by the Vidal lab (29), in which Y2H screens were conducted in parallel against both cDNA and ORFeome libraries and then compared for the protein interactions identified. In total, 1,517 interactions were identified with the cDNA library and 3,263 with the ORFeome library. Only 250 interactions were discovered using both libraries, thus representing 5.5% overlap. Furthermore, the viral polymerase genes used as baits came from different influenza virus strains compared to the results from Shapira et al. (51) and our I-MAP data set.
Table 2.
Overlap in FluPol interactors between different data sets
| Data set | No. of HFlu proteins | No. (%) of interactors with overlapa: |
|||
|---|---|---|---|---|---|
| Y2H, I-MAP | Y2H, Shapira et al. (51) | LC | AP | ||
| Y2H,I-MAP | 109 | 1 (0.9) | 3 (2.8) | 2 (1.8) | |
| Y2H, Shapira et al. (51) | 69 | 1 (1.4) | 2 (2.9) | 0 | |
| LC | 23 | 3 (13.0) | 2 (8.7) | 8 (34.8) | |
| AP | 49 | 2 (4.1) | 0 | 8 (16.3) | |
The overlap is represented as percentage of the total HFlu data set analyzed (horizontally). For example, the overlap of 1 interactor between Y2H I-MAP and Y2H Shapira et al. corresponds to 0.9% of the 109 HFlu proteins identified in Y2H I-MAP.
The FluPol interactome we reconstructed is thus composed of 4 viral proteins and 234 human partners (HFlu proteins) forming 279 viral-human protein interactions (Fig. 1). For a dynamic version of the network, see http://flupol.lyon.inserm.fr and Table S1 in the supplemental material). Using VirHostNet (37), we found that among the 234 HFlu proteins, 111 are connected with each other through 204 interactions within the human interactome (HFlu-HFlu) (Fig. 1; see Table S2 in the supplemental material). We also found through VirHostNet that 69 HFlu proteins are targeted by 47 other viruses (see Table S3 in the supplemental material). This suggests that cellular pathways that are connected to the influenza virus polymerase (see below) represent common virus targets. Furthermore, 24 influenza virus polymerase interactors are also connected to other influenza A virus proteins (mainly NS1; see Table S4 in the supplemental material), centering the polymerase partners in a more comprehensive influenza virus network.
Functional enrichment of the influenza virus polymerase interactome.
To visualize which cellular functions are targeted by the viral polymerase, the enrichment of HFlu for Gene Ontology (GO) terms corresponding to “biological processes” was determined by using the BiNGO plugin in Cytoscape (1, 16, 32, 50). It is biased, since not all human proteins have yet been annotated (in our data set, 208 of the 234 HFlu proteins possess a GO “biological process” annotation), but it remains a powerful way to incorporate conventional biology to system-level data sets. We took into account the false-positive rate obtained during the validation of the Y2H interactors and performed 50 tests by removing randomly 15% of the Y2H cellular partners. We retained only GO terms that were significantly enriched in all 50 tests. Functions related to DNA synthesis (DNA replication and DNA repair), transcription, RNA processing, and translation were overrepresented. An enrichment for cell cycle, nucleocytoplasmic transport, response to unfolded proteins, and viral infectious cycle was also found (Fig. 1C). Among these overrepresented processes, several were mainly retrieved from literature—e.g., nucleocytoplasmic transport (10/14 cellular interactors were mined from literature) or DNA replication (6/10 interactors from literature)—denoting functions already known to be associated with the influenza virus polymerase (11, 13, 25, 34, 35, 39, 46, 57). Other processes contain several interactors discovered in our Y2H screens, like RNA processing (11/27 proteins identified from Y2H assay), DNA repair (8/18 proteins identified from Y2H assay), unraveling physical links between the influenza virus polymerase and these cellular functions. They may be mandatory for the function of influenza virus polymerase within the host cell.
Functional implication of the FluPol partners.
On the basis of the enriched cellular functions that are targeted by the influenza virus polymerase, we selected eight validated cellular interactors to test their ability to affect the viral replication: C14orf166 (not annotated in GO but known as a transcription elongation factor) (20, 44), COPS5 (implicated in transcription and in translation), EIF3S6IP (translation), FANCG (DNA repair), MNAT1 (a member of 4 overrepresented classes, including cell cycle, RNA processing, transcription, and DNA repair), NMI (transcription), NUP54 (nucleocytoplasmic transport), and PRKRA (RNA processing and also implicated in innate antiviral response and known to interact with NS1) (our unpublished data and reference 39) (Fig. 1C).
To further confirm observed interactions, we tested whether these cellular partners interact with the viral polymerase in influenza virus-infected cells. For this purpose, we transfected cells with each protein fused to the 3× Flag epitope and then infected them with H1N1 A/New Caledonia/2006 virus (clinical isolate). Twenty-four hours after infection, Flag-tagged cellular proteins were immunoprecipitated, and Western blots were performed with an antibody raised against PB1 to detect the presence of the polymerase in the immunoprecipitates. As shown in Fig. 2, COPS5, EIF3S6IP, FANCG, NMI, NUP54, and PRKRA immunoprecipitated the viral polymerase (detection of PB1 in the immunoprecipitates). We did not detect PB1 in the C14orf166 and MNAT1 immunoprecipitates. Nevertheless, we noticed that C14orf166 interacts with PA but not with the other polymerase subunits (Table 1). This might reflect that C14orf166 only interacts with free PA protein. In summary, these results indicate that 6 of the 8 host cell factors tested specifically associate with the RdRP in an infected cell context.
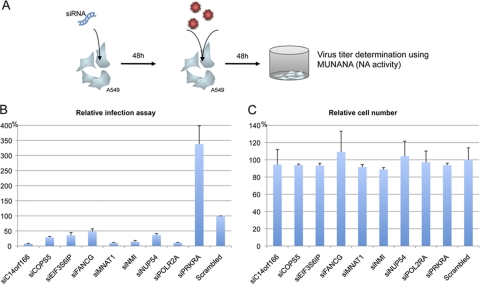
We then monitored the virus replication in human cells where these 8 influenza virus polymerase interactors were depleted by specific siRNA. We added to this set POLR2A, which we identified by Y2H assay and which was previously described in the literature as an RdRP interactor (14). For this purpose, siRNA-depleted cells were infected by H1N1 virus, and 48 h after infection, virus titers were determined by quantifying the neuraminidase activity using a MUNANA assay (59) (see Materials and Methods). The individual depletion of 8 genes decreased the virus replication to less than 50% compared to a control scrambled siRNA: C14orf166, COPS5, EIF3S6IP, FANCG, MNAT1, NMI, NUP54, and POLR2A (Fig. 3B). Conversely, PRKRA appeared to have potent antiviral activity, as its inhibition by siRNA increased the virus replication 3.5 times compared to the control scrambled siRNA. As a control, we showed that tested siRNA had no side effects on cell survival as assessed by a resazurin assay (Fig. 3C).
Fig. 3.
FluPol-host cell interactors regulate virus replication. (A) Schematic representation of the replication assay. Human A549 cells treated with targeted siRNA were infected 48 h after depletion by H1N1 A/New Caledonia/2006 at an MOI of 0.5. Cell culture supernatants were harvested 48 h after infection, and viral titers were determined by a MUNANA assay (measuring the neuraminidase [NA] activity). (B) Results (mean of 2 independent experiments, each performed in triplicate) are expressed as relative replication efficiency compared to cells treated with a scrambled siRNA control. (C) To assess potential cytotoxicity induced by the siRNAs, a cell viability assay was performed using the resazurin-based fluorometric assay 48 h after siRNA transfection. Cells were counted and expressed as relative to control cells (treated with a scrambled siRNA control).
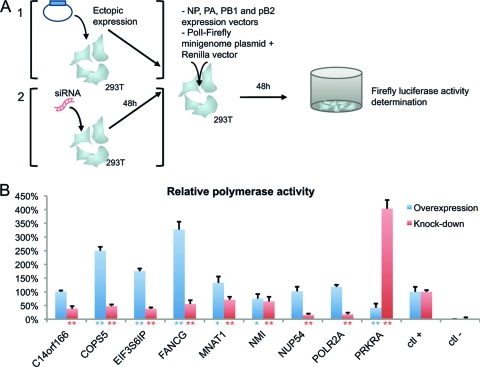
We speculated that these cellular proteins specifically regulate the viral polymerase activity. This was tested in a minigenome replicon assay either by overexpressing (by transient transfection of a plasmid) or by depleting (using directed siRNA) each cellular partner gene. HEK-293T cells were cotransfected with vectors encoding the A/WSN/33 polymerase subunits and NP and a reporter plasmid encoding an RNA template for the viral polymerase that expresses the firefly luciferase, together with a plasmid expressing each cellular partner. Luciferase activity was determined 48 h after transfection. For the depletion assay, human cells were first treated by siRNA targeting each gene for 2 days and then were transfected with the complete set of the vRNP (plasmids expressing NP, PA, PB1, and PB2 and the reporter plasmid). Results are presented in Fig. 4. The eight genes required for virus replication reveal an increased polymerase activity when overexpressed and/or a decreased activity when depleted. COPS5, EIF3S6IP, FANCG, and MNAT1 overexpression enhanced the RdRP transcriptional activity from 1.25 to 3 times compared to the control. The depletion of C14orf166, COPS5, EIF3S6IP, NUP54, and POLR2A reduced more than 2 times the viral polymerase transcription. We did not observe a drastic change in the RdRP transcription when overexpressing or depleting NMI. This cellular protein could contribute to viral replication at specific steps that cannot be detected with the minigenome system, like virus assembly or trafficking (2, 62). These human proteins are thus host cell factors required for influenza virus polymerase activity, a mandatory step for influenza virus replication. On the contrary, PRKRA acts as an antiviral cellular factor, since its overexpression inhibits the viral polymerase activity and its knockdown favors the influenza virus polymerase transcriptional activity as well as the virus replication.
Fig. 4.
FluPol-host cell interactors regulate the viral polymerase activity. (A) Schematic representation of the viral polymerase transcriptional activity using a minigenome replicon assay. Human 293T cells were either transfected by a vector encoding the cellular interactor (section 1, overexpression test) or treated with a siRNA targeting the cellular gene (section 2, depletion test). For the overexpression assay (in blue), 293T cells were transfected with a plasmid encoding each cellular protein together with NP, PA, PB1, PB2, a minigenome replicon reporter plasmid coding for firefly luciferase, and a Renilla control vector. Forty-eight hours posttransfection, the luciferase activities were determined. For the depletion assay (in red), 293T cells were first treated with specific siRNAs targeting cellular partners' genes. Forty-eight hours after depletion, cells were transfected with the complete set of vRNP (NP, PA, PB1, and PB2 and the luciferase reporter plasmid) and the Renilla control vector, and the experiment proceeded as for the overexpression test. (B) Results (mean of 2 independent experiments, each performed in triplicate) are expressed as relative effect of overexpression (in blue) or knockdown of FluPol interactors (in red) compared to the control (empty vector for overexpression or scrambled siRNA for depletion). As a negative control (ctl −), cells were transfected with the same reaction mixture without PB1. * and **, P < 0.05 and P < 0.01, respectively, compared to the positive control (ctl +), based on a Student's t test.
Among the eight proviral cellular proteins, 5 have been implicated in mRNA transcription: C14orf166, COPS5, MNAT1, NMI, and POLR2A. The latter protein, the largest polymerase II (Pol II) subunit, interacts with the viral polymerase through its carboxy-terminal domain (CTD) phosphorylated at serine 5 (i.e., when the transcription initiates and where the capping enzymes are recruited and activated) (14). This interaction inhibits the transcription elongation and leads to the degradation of POLR2A (6, 47). We noticed in a very recent paper that influenza virus polymerase interacts with the P-TEFb complex (CDK9/cyclin T1), which phosphorylates the Pol II CTD at the onset of transcription elongation (61). This further connects the influenza virus polymerase complex to the RNA Pol II at the beginning of the transcription elongation (i.e., when cellular pre-mRNAs are capped) (reviewed in reference 8). We have shown that the POLR2A CTD specifically and directly interacts with PA: the clones isolated in the Y2H assay all correspond to the POLR2A CTD, and in the FCPI assay, it interacted only with PA (and not with NP, PB1, or PB2) (Table 1). In biochemical fractionation experiments, the actively transcribing Pol II form and many proteins involved in cellular transcription and RNA processing are associated with an insoluble nuclear fraction, the nuclear matrix (36). Interestingly, newly synthesized viral RNA and components of the vRNP complex have been found associated with this nuclear matrix in influenza virus-infected cells, and protein interactions identified in this study probably participate in this process (14, 21).
PRKRA, which we identified as a protein required for the host antiviral response, is a double-stranded RNA (dsRNA) binding protein that activates protein kinase R (PKR) by interacting through their dsRNA binding domains (41). PKR, a key mediator of the interferon antiviral pathway, is a kinase that phosphorylates the translation initiation factor eIF2α, leading to protein synthesis shutoff (7). The overexpression of PRKRA in mammalian cells leads to PKR activation and inhibition of translation and, under cellular stress, to apoptosis (40, 41). It has already been shown that PRKRA interacts with viral proteins, as the dsRNA binding protein Us11 of HSV1. This interaction inhibits PKR activation and the PRKRA-induced apoptosis (45). Recently, vesicular stomatitis virus (VSV) replication was monitored in a PRKRA-dependent context, showing that the virus production is enhanced in PRKRA-depleted cells and that PRKRA overexpression protects cells against VSV infection (3), like we observed in influenza virus replication assay. Furthermore, it has also been shown that the influenza virus NS1 protein interacts with both PRKRA and PKR, leading to the inhibition of PKR and stress-induced cell death mediated by PRKRA (30). It appears thus that the PRKRA-PKR pathway is very specifically targeted by the influenza virus, and this might be an important way for the virus to escape the innate immune response in infected cells.
In their comprehensive influenza virus infection regulatory study, Shapira and colleagues identified the viral polymerase subunits as modulating factors of the host antiviral response, like the interferon production pathway (51). Furthermore, influenza virus RdRP was recently shown to interact with Stau1, a dsRNA binding protein and well-characterized target of NS1 (10).
This global approach paves the way to a new landscape of the influenza virus polymerase cellular neighborhood. The influenza virus polymerase interaction network will help us to better decipher the virus biology and molecular mechanisms of viral replication. By adding a functional correlation between the influenza virus polymerase and cellular cofactors, it will facilitate the development of new antiviral drugs that target the host cell factors instead of viral proteins, the latter being more prone to mutate and become drug resistant.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Hart (H3N2 A/Hong Kong/43/75 PB2 ORF), G. Brownlee (H1N1 A/WSN/33 ORFs encoding vectors), V. Moulès (H1N1 A/PR/8/34 genomic cDNA, H5N1 A/VietNam/1194/2004 and H5N1 A/Turkey/651242/2006 genomic RNA), J. Ortin (polyclonal antibody against PB1), Y. Jacob (pCMV-eGFP-gw and pCI-neo-3Flag-gw vectors), and M. Shaw (minigenome reporter plasmid) for providing materials. We also thank all of the members of the I-MAP team for fruitful discussions.
This work was supported by the EU FLUPOL (SP5B-CT-2007-044263), ANR FLU INTERPOL (ANR-06-MIME-014-02), the French Fonds Unique Interministériel (FUI), and INSERM.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 12 October 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Ashburner M., et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bao J., Zervos A. S. 1996. Isolation and characterization of Nmi, a novel partner of Myc proteins. Oncogene 12: 2171–2176 [PubMed] [Google Scholar]
- 3. Bennett R. L., et al. 2006. RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy, and viral infection. Blood 108: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blazer L. L., Roman D. L., Muxlow M. R., Neubig R. R. 2010. Use of flow cytometric methods to quantify protein-protein interactions. Curr. Protoc. Cytom. 51: 13.11.1–1311.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brass A. L., et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan A. Y., Vreede F. T., Smith M., Engelhardt O. G., Fodor E. 2006. Influenza virus inhibits RNA polymerase II elongation. Virology 351: 210–217 [DOI] [PubMed] [Google Scholar]
- 7. Colthurst D. R., Campbell D. G., Proud C. G. 1987. Structure and regulation of eukaryotic initiation factor eIF-2. Sequence of the site in the alpha subunit phosphorylated by the haem-controlled repressor and by the double-stranded RNA-activated inhibitor. Eur. J. Biochem. 166: 357–363 [DOI] [PubMed] [Google Scholar]
- 8. de Almeida S. F., Carmo-Fonseca M. 2008. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 582: 1971–1976 [DOI] [PubMed] [Google Scholar]
- 9. de Chassey B., et al. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Lucas S., Peredo J., Marion R. M., Sanchez C., Ortin J. 2010. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. J. Virol. 84: 7603–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng T., et al. 2006. Role of Ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80: 11911–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dias A., et al. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914–918 [DOI] [PubMed] [Google Scholar]
- 13. Elton D., et al. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75: 408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engelhardt O. G., Smith M., Fodor E. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79: 5812–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finkelstein D. B., et al. 2007. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 81: 10292–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia O., et al. 2007. GOlorize: a Cytoscape plug-in for network visualization with Gene Ontology-based layout and coloring. Bioinformatics 23: 394–396 [DOI] [PubMed] [Google Scholar]
- 17. Guilligay D., et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15: 500–506 [DOI] [PubMed] [Google Scholar]
- 18. Hao L., et al. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454: 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffmann H. H., Palese P., Shaw M. L. 2008. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antiviral Res. 80: 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huarte M., Sanz-Ezquerro J. J., Roncal F., Ortin J., Nieto A. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75: 8597–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson D. A., Caton A. J., McCready S. J., Cook P. R. 1982. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 296: 366–368 [DOI] [PubMed] [Google Scholar]
- 22. Jorba N., Coloma R., Ortin J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5: e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jorba N., et al. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8: 2077–2088 [DOI] [PubMed] [Google Scholar]
- 24. Karlas A., et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463: 818–822 [DOI] [PubMed] [Google Scholar]
- 25. Kawaguchi A., Nagata K. 2007. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26: 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerppola T. K. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konig R., et al. 2010. Human host factors required for influenza virus replication. Nature 463: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krug R. M., Broni B. A., Bouloy M. 1979. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 18: 329–334 [DOI] [PubMed] [Google Scholar]
- 29. Li S., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li S., Min J. Y., Krug R. M., Sen G. C. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349: 13–21 [DOI] [PubMed] [Google Scholar]
- 31. Lopes C. T., et al. 2010. Cytoscape Web: an interactive web-based network browser. Bioinformatics 26: 2347–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maere S., Heymans K., Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- 33. Mayer D., et al. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6: 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Melen K., et al. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278: 28193–28200 [DOI] [PubMed] [Google Scholar]
- 35. Momose F., et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75: 1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mortillaro M. J., et al. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. U. S. A. 93: 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Navratil V., et al. 2009. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 37: D661–D668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neumann G., Brownlee G. G., Fodor E., Kawaoka Y. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 283: 121–143 [DOI] [PubMed] [Google Scholar]
- 39. O'Neill R. E., Palese P. 1995. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology 206: 116–125 [DOI] [PubMed] [Google Scholar]
- 40. Patel C. V., Handy I., Goldsmith T., Patel R. C. 2000. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275: 37993–37998 [DOI] [PubMed] [Google Scholar]
- 41. Patel R. C., Sen G. C. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17: 4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pellet J., et al. 2009. pISTil: a pipeline for yeast two-hybrid Interaction Sequence Tags identification and analysis. BMC Res. Notes 2: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pellet J., et al. 2010. ViralORFeome: an integrated database to generate a versatile collection of viral ORFs. Nucleic Acids Res. 38: D371–D378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez-Gonzalez A., Rodriguez A., Huarte M., Salanueva I. J., Nieto A. 2006. hCLE/CGI-99, a human protein that interacts with the influenza virus polymerase, is a mRNA transcription modulator. J. Mol. Biol. 362: 887–900 [DOI] [PubMed] [Google Scholar]
- 45. Peters G. A., Khoo D., Mohr I., Sen G. C. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76: 11054–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Resa-Infante P., et al. 2008. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3: e3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez A., Perez-Gonzalez A., Nieto A. 2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 81: 5315–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rual J. F., et al. 2004. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14: 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salomon R., et al. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203: 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shannon P., et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shapira S. D., et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Subbarao E. K., London W., Murphy B. R. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67: 1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tafforeau L., Rabourdin-Combe C., Lotteau V. Virus-human cell interactomes. Methods Mol. Biol., in press [DOI] [PubMed] [Google Scholar]
- 54. Tarendeau F., et al. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14: 229–233 [DOI] [PubMed] [Google Scholar]
- 55. Tarendeau F., et al. 2008. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 4: e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vreede F. T., Jung T. E., Brownlee G. G. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78: 9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang P., Palese P., O'Neill R. E. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71: 1850–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Watanabe T., Watanabe S., Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yongkiettrakul S., et al. 2009. Avian influenza A/H5N1 neuraminidase expressed in yeast with a functional head domain. J. Virol. Methods 156: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yumerefendi H., Tarendeau F., Mas P. J., Hart D. J. 2010. ESPRIT: an automated, library-based method for mapping and soluble expression of protein domains from challenging targets. J. Struct. Biol. 172: 66–74 [DOI] [PubMed] [Google Scholar]
- 61. Zhang J., Li G., Ye X. 2010. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J. Virol. 84: 12619–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhu M., John S., Berg M., Leonard W. J. 1999. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell 96: 121–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.