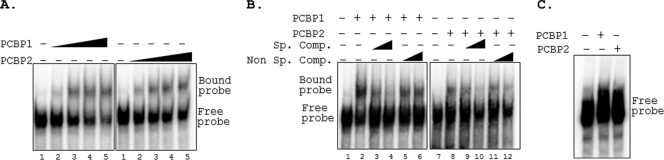
Fig. 4.
PCBP1 and PCBP2 form complexes with PRRSV 5′UTR. (A) EMSA of 32P-labeled PRRSV (+) 5′UTR probe in the presence of increasing concentrations (0, 0.5, 0.75, 1, and 1.25 μg in lanes 1 to 5) of recombinant purified PCBP1 or PCBP2 in an RNA binding reaction. RNP complexes were resolved by 5% nondenaturing PAGE and visualized by exposing the dried gel to phosphorimager screens. The positions of the free and bound probes are indicated on the right. (B) Competition gel mobility shift assay. Radiolabeled PRRSV (+) 5′UTR probes were incubated with 0.75 μg of PCBP1 or PCBP2 in presence (+) or absence (−) of unlabeled 5′UTR probe (Sp. comp.) or an irrelevant probe (Non Sp. comp.). Lanes: 1 and 7, free probe; 2 and 8, no competitor; 3, 4, 9, and 10, increasing concentration of specific competitor (10-fold and 50-fold molar excess); 5, 6, 11, and 12, increasing concentration of nonspecific competitor (10-fold and 50-fold molar excess). (C) Gel mobility shift assay with 3′UTR. Radiolabeled PRRSV (+) 3′UTR probe was incubated without (−) or with (+) 1 μg of PCBP1 or PCBP2, and EMSA was performed as described above.