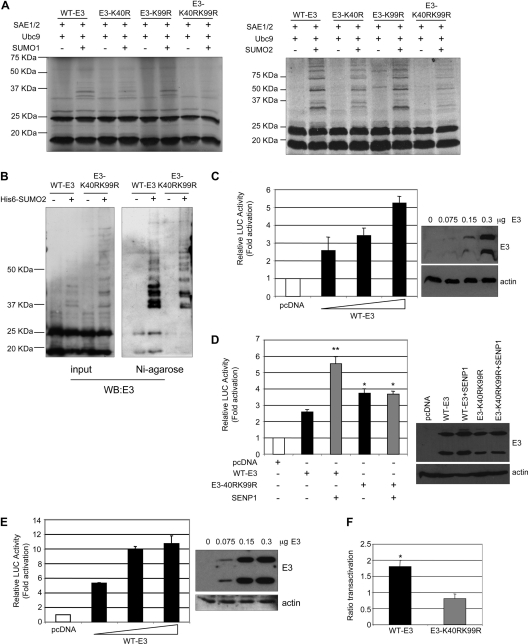
Fig. 3.
Role of E3 SUMOylation in transcriptional activity. (A) [35S]methionine-labeled wild-type or mutant E3 proteins were used as substrates in an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2. The reaction products were visualized by autoradiography. (B) Total extracts or histidine-tagged purified proteins were prepared from HEK-293 cells cotransfected with pCINEO-E3-WT or E3-K40RK99R together with pcDNA or pcDNA-Ubc9 and pcDNA-His6-SUMO2, and an immunoblot analysis with an anti-E3 antibody was carried out. (C and D) MCF-7 cells were transiently transfected in triplicate with the luciferase reporter PUMA-luc together with the plasmids indicated. The same results were obtained in at least three different experiments. Data represent means ± standard errors (SE) for one experiment. *, P < 0.05; **, P < 0.005 (compared with WT-E3-transfected cells; determined by the Student test). The right panel represents the expression levels of the E3L gene transfected in each experiment. (E) MCF-7 cells were transiently transfected in triplicate with the luciferase reporter APAF-luc together with the plasmids indicated. The same results were obtained in at least three different experiments. Data represent means ± SE for one experiment. (F) Ratio of the level of transactivation of APAF-1–luc by WT-E3 or E3-K40RK99R and SENP-1 to the level of transactivation by WT-E3 or E3-K40RK99R alone, respectively. The same results were obtained in at least three different experiments. Data represent means ± SE for one experiment. *, P < 0.05 (compared with WT-E3 transfected cells; determined by the Student test).