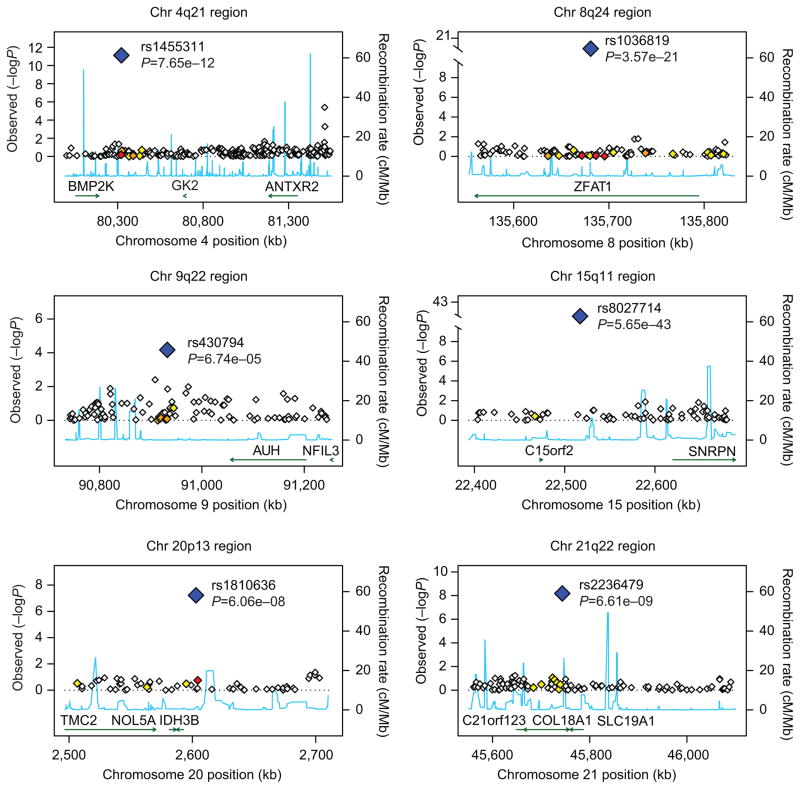
Figure 3.
Plots of the six chromosomal regions found to be of interest. Gene plots are based on NCBI35/hg17. Estimated underlying recombination rates for a region are based on HapMap CEU individuals and are shown in blue. Markers in high linkage disequilibrium with one of the six single nucleotide polymorphisms (SNPs) of interest are determined using r-squared values obtained from HapMap CEU and are coded by the color of the diamond shape. White indicates no linkage disequilibrium (r2<0.2), yellow indicates weak linkage disequilibrium (r2 between 0.2 and 0.5), orange indicates moderate linkage disequilibrium (r2 between 0.5 and 0.8), and red indicates strong linkage disequilibrium (r2 0.8 or greater).