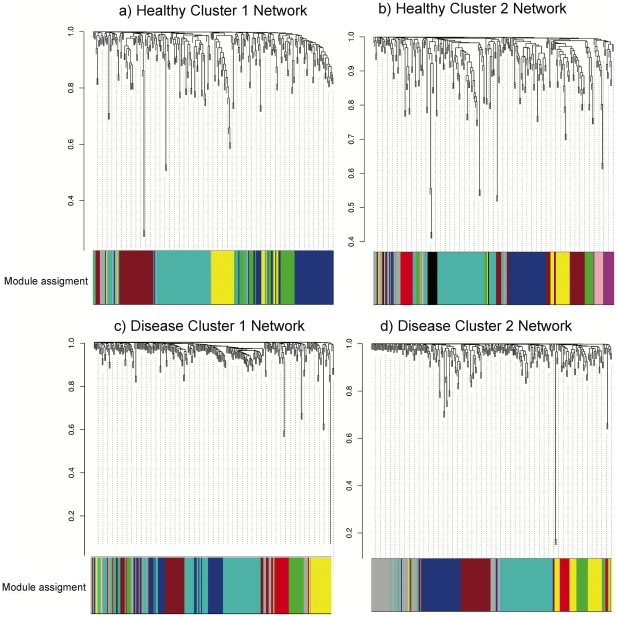
Figure 2. WGCNA correlation network results of bacterial species in healthy and diseased individuals from HOMIM results.
Clustering dendrogram of species, with dissimilarity based on topological overlap, together with assigned module colors. a) Cluster 1 from healthy individuals (51 samples), R2 used for scale free topology model fit was 0.90 and a total of 6 bacterial modules were identified. b) Cluster 2 from healthy individuals (37 samples), R2 used for scale free topology model fit was 0.85 and a total of 10 bacterial modules were identified. c) Cluster 1 from diseased individuals (467 samples), R2 used for scale free topology model fit was 0.90 and a total of 6 bacterial modules were identified. D) Cluster 2 from diseased individuals (49 samples), R2 used for scale free topology model fit was 0.85 and a total of 7 bacterial modules were identified.