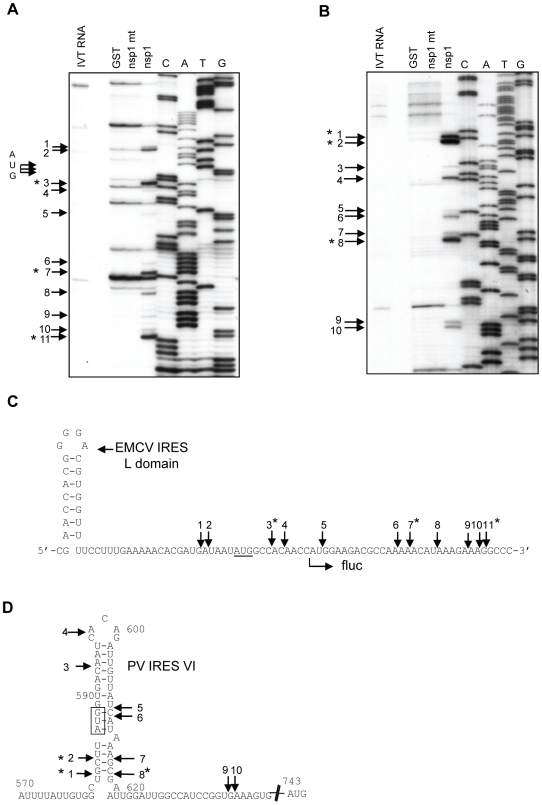
Figure 3. Primer extension analysis of Ren-EMCV-FF and Ren-PV-FF after incubation with GST, nsp1 or nsp1-mt.
(A, B) In vitro-synthesized Ren-EMCV-FF (A) and Ren-PV-FF (B) were incubated with GST, nsp1 or nsp1-mt in RRL (A) and RRL+HeLa (B), respectively. The RNAs were extracted and subjected to primer extension analysis by using the 5′-end 32P labeled primer that binds at ∼100 nt downstream of the fluc translation initiation codon for Ren-EMCV-FF (A) or a labeled primer that binds 6 nt downstream of the fluc initiation codon region for Ren-PV-FF (B). Primer extension products and DNA sequence ladders, which were generated from the plasmid used for RNA synthesis and the primer used for primer extension analysis, were resolved on 8% polyacrylamide/7M Urea DNA sequencing gels. Premature primer extension termination signals are indicated by enumerated arrows and major RNA modification sites are marked with asterisks. IVT RNA, primer extension analysis of in vitro-transcribed RNAs that were not incubated in RRL; AUG, translation initiation AUG (AUG-834) of EMCV IRES. (C, D) RNA modification sites in Ren-EMCV-FF (C) and Ren-PV-FF (D). The structural domain L of the EMCV IRES (C) [39] in Ren-EMCV-FF and the poliovirus IRES stem-loop IV (D) [75] in Ren-PV-FF are also shown. Arrows indicate RNA modification sites and asterisks denote the main ones. The underlined AUG triplet is equivalent to the translation initiation codon AUG-834 in the EMCV genome (C) and the boxed AUG triplet is equivalent to the AUG-586 of the poliovirus genome. AUG-586 of the poliovirus is mapped ∼150 nt upstream of the authentic viral translation initiation codon.