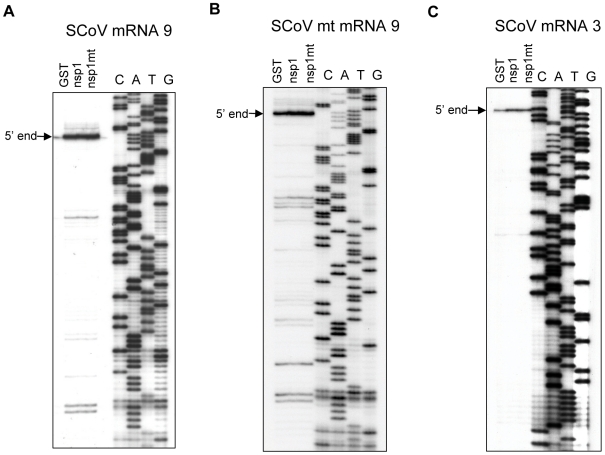
Figure 9. Susceptibilities of naturally occurring SCoV mRNAs to nsp1-induced RNA modification.
(A and C) Poly A+ RNA was extracted from SCoV-infected cells and incubated with GST, nsp1 or nsp1-mt in RRL. Extracted RNAs were subjected to primer extension analysis by using primers suitable for detecting RNA modifications in SCoV mRNA 9 (A) or in SCoV mRNA 3 (C). Primer extension products were resolved in 8% polyacrylamide/7M Urea DNA sequencing gels: 5′-end, full-length primer extension product of SCoV mRNA 9 (A) or SCoV mRNA 3 (C). (B) Similar experiments described in (A) were performed, except that RNAs from SCoV-mt-infected cells were used.