Figure 1.
Muscle-Specific lincRNA Profiling
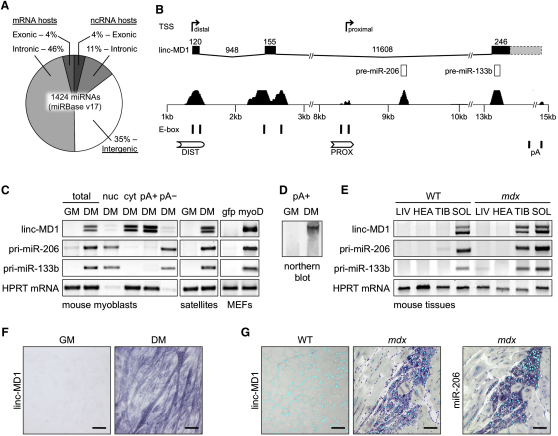
(A) Human miRNA genomic location relative to their host genes.
(B) Schematic representation of the murine miR-206/133b genomic locus. Upper panel shows transcriptional start sites (TSS, indicated by arrows) mapped through 5′ RACE analysis in differentiated myoblasts. The genomic structure of the identified linc-MD1 and the exon-intron lengths are shown (conserved exons in black, nonconserved 3′-portions in dash) as well as pre-miRNA sequences. Lower panel shows conserved regions among vertebrates together with E-boxes and regulatory elements (DIST, PROX and pA signals).
(C) RT-PCR for linc-MD1, pri-miR-206, and pri-miR-133b expression performed in mouse myoblasts in growth (GM) or differentiation (DM) conditions. The total, nuclear (nuc), cytoplasmic (cyt), polyadenylated (pA+) and nonpolyadenylated (pA-) RNA fractions are shown. The same analysis was also performed in primary satellite cells in GM and DM conditions and in mouse embryonic fibroblasts (MEFs) infected with a MyoD expressing lentivirus (MyoD) or control (gfp). HPRT mRNA was used as endogenous control.
(D) Northern blot analysis for linc-MD1 in the poly-A+ fraction from mouse myoblasts in GM and DM conditions.
(E) RT-PCR for linc-MD1, pri-miR-206, pri-miR-133b performed on RNA from liver (LIV), heart (HEA), tibialis anterior (TIB) and soleus (SOL) of wild-type (WT) and mdx mice.
(F) In situ analysis with a DIG-labeled linc-MD1 probe in C2 myoblasts in GM and DM. (G) In situ analysis with DIG-labeled linc-MD1 and miR-206 probes in wild-type (WT) and mdx gastrocnemius cryosections; DAPI staining (light blue) is also shown.
Original magnification is 20×. The scale bar represents 100 μm. Additional fields and controls are show in Figure S2.