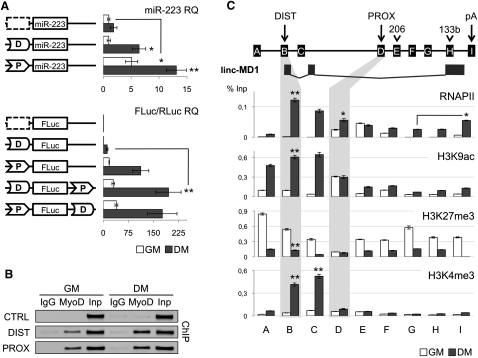
Figure 2.
Transcriptional and Epigenetic Regulation of miR-206/133b Genomic Locus
(A) Promoter activity assay. Upper panel shows that distal (box D) and proximal (box P) regions were cloned upstream of murine pre-miR-223 and tested in C2 myoblasts in GM and DM conditions. A region between miR-206 and miR-133b was used as negative control (dashed box). miR-223 expression was measured by northern blot (Figure S3) and qRT-PCR. U6 snRNA was used as endogenous control. miR-223 relative quantification (RQ) is shown with respect to control vector in GM set to a value of 1. Lower panel shows that the same regions were cloned, alone or in combinations, in a firefly luciferase reporter construct (FLuc) and tested in GM and DM conditions. A luciferase construct (RLuc) was transfected as control. Luciferase activity, from three independent experiments, was measured as FLuc/RLuc RQ shown with respect to the negative control vector in GM set to a value of 1.
(B) ChIP analysis for MyoD enrichment on distal (DIST), proximal (PROX) and negative control (CTRL) regions performed on chromatin extracted from C2 myoblasts in GM and DM conditions. Amplifications of IgG control immunoprecipitations (IgG) and 10% of input chromatin (Inp) are shown.
(C) Upper panel is a schematic representation of the miR-206/133b genomic locus. Capital letters indicate regions analyzed in ChIP experiments. Location of regulatory regions (DIST, PROX and pA), pre-miRNAs (206, 133b) and linc-MD1 are shown along the locus as in Figure 1B. Lower panel displays ChIP analyses for RNA polymerase II (RNAP II), H3K9ac, H3K27me3, and H3K4me3 performed on chromatin extracted from myoblasts in GM and DM. Values derived from three independent experiments were normalized for background signals (IgG) and expressed as percentage of Input chromatin (% Inp). Unless specifically indicated, statistical significance was calculated with respect to GM conditions. Data are shown as mean ± SD. One asterisk indicates p < 0.05; two asterisks: p < 0.01.